Abstract
Epithelial-to-mesenchymal transition (EMT) as well as the reverse process, mesenchymal-to-epithelial transition (MET) is important during embryogenesis. EMT is also involved in cancer invasion and metastasis, and can generate cells with properties similar to those of stem cells. Retrotransposons can rearrange the genome by inserting DNA in new loci, thus inducing mutations. This study examines the gene expression of transcription factors involved in EMT and MET. In the second experimental panel, the gene expression of L1 retrotransposon was studied. L1-open reading frame (ORF) 2 mRNA was found to be expressed both in cancer and cancer stem cells, while L1-ORF1 mRNA was expressed only in cancer cells. The suppression of L1-ORF2 gene expression demonstrated that this retrotransposon might affect EMT in colon cancer stem cells. This study highlights that the EMT process seems to differ between cancer cells and cancer stem cells, and that transposable elements seem to be involved in the process, influencing cellular plasticity.
INTRODUCTION
Cellular plasticity refers to the ability of cells to take on characteristics of other types of cells, in particular those of stem cells. Epithelial-to-mesenchymal transition (EMT) is a biological process that triggers epithelial cells to assume a mesenchymal cell phenotype through interactions at the basement membrane and multiple biochemical changes. EMT is involved in mesoderm formation, wound healing, and other cell and organ development processes; however, it is also important for the initiation of metastasis (CitationKalluri, 2009). Epithelial cancer cells can migrate through the basement membrane into the bloodstream and become circulating tumor cells (CTCs). These cells can then exit the bloodstream and initiate the process opposite to EMT, mesenchymal- to-epithelial transition (MET), to form micrometastases (CitationChaffer, 2011). The loss of E-cadherin expression is the first indication that a cell is undergoing EMT, followed by an increase in expression of the mesenchymal markers, N-cadherin, fibronectin, and vimentin. Studies have identified many signal transduction pathways that are involved in the mediation and regulation of EMT (CitationLee, 2006). Recently, a sub-population of CTCs has been found to possess tumorigenic properties, and these have been dubbed cancer stem cells (CSCs) (CitationToloudi, 2011). Cells that undergo EMT gain stem cell properties, thus connecting both EMT and CSCs to metastasis and cancer progression. However, a clear understanding of which factors activate the specific transcription factors that regulate the stemness, epithelial, and mesenchymal phenotypes remains elusive. Given that approximately 45% of the human genome is made up of transposons, particularly retrotransposons (42%), we questioned whether these elements are involved in EMT and MET (CitationLander, 2001). The L1 retrotransposon consists of two different open reading frames (ORFs), each one with particular properties. The ORF1p has nucleic acid chaperone activity, while ORF2p is responsible for retrotransposition, as it contains both endonuclease and reverse transcriptase activities (CitationFeng, 1996). The current study first examines the presence and expression of long interspersed element 1 (LINE-1) mRNA in a wide spectrum of cancer, cancer stem, and normal cells. We then explore the role of L1 in the plasticity and stemness of colon CSCs by studying the gene expression of many transcription factors involved in both processes.
METHODS AND MATERIALS
Cell culture
Human colon CSCs (36112-39P; Celprogen), breast CSCs (36102-29P; Celprogen), and lung CSCs (36107-34; Celprogen) were cultured in appropriate growth medium (M36112-39U, M36102-29U, and M36107-34U, respectively; Celprogen), supplemented with 10% fetal bovine serum (FBS; 10270-106; Gibco) in 25 cm2 flasks (E36102-29P-T25, E36102-29P-T25, E36107-34-T25; Celprogen) at 37°C in a 5% CO2 environment. The human HCT-116 colon carcinoma cells (91091005; ECACC), and T47D (85102201; ECACC) breast tumor cells were cultured in Dulbecco's modified Eagle's medium (D5546; Sigma Aldrich). Finally, the human lung cancer cell line COLO699N (93052608; ECACC) was cultured in RPMI-1640 medium (R0883; Sigma Aldrich), supplemented with 10% fetal bovine serum (FBS) and L-glutamine (G5792; Sigma Aldrich) in 25 cm2 flasks (430639; Corning) under the same conditions. Peripheral blood mononuclear cells (PBMCs) were obtained from a healthy 30-year-old male. Twenty milliliters of peripheral blood was collected from the donor and placed in a tube with ethylenediaminetetraacetic acid (EDTA) as anticoagulant (Vacutainer K3E; BD) and rotated for 30 min to prevent coagulation. To determine the presence of cancer cells in our sample, flow cytometry was performed as a method of choice. To isolate PBMCs, whole blood cells were centrifuged with polysucrose solution (Biocoll separating solution 1077; Biochrom). Mononuclear cells, lymphocytes, platelets, and granulocytes were collected after centrifugation and washed twice with phosphate-buffered saline (PBS). Cells were then incubated at 4°C for 30 min with EpCAM magnetic beads (39-EPC-50; Gentaur). EpCAM is an epithelial cell adhesion molecule-associated antigen that is expressed in the vast majority of carcinomas (CitationMunz, 2009). Following incubation, the sample was placed in a magnetic field, selected, and washed with PBS. The negative selection cells (non-cancerous) were isolated and then cultured in a 25-cm2 flask (5520100; Orange Scientific) with RPMI-1640 medium (R6504; Sigma).
Evaluation of cells
Cellular assays were based on the ability of CSCs to form microspheres in semi-suspension, observed with an inverted light microscope. The cultures used have previously been evaluated by molecular analyses, including gene expression analysis for specific transcription factors (CitationChiou, 2008; CitationXiang, 2011; CitationJanikova, 2010). The authenticity of the control cell line was tested using molecular assays including short tandem repeat (STR) profiling, the results of which were compared with the manufacturer's profile. Cultivation was continued for more than 30 passages to exclude the possibility of incorporating embryonic stem cells (ESCs) in the experiments, since CSCs are immortal unlike ESCs. STRs analysis was also performed for the normal sample in each passage, to ensure the stability of genotype. Furthermore, the stability of the phenotype was tested in each passage, according to the manufacturer's patterns.
Molecular analysis
DNA and RNA were extracted from cell cultures using the QIAamp DNA Mini Kit (51306; Qiagen) and RNeasy Mini Kit (74105; Qiagen), respectively. Both samples were evaluated spectrophotometrically, with the RNA samples also subjected to agarose gel electrophoresis to verify integrity of the 18S–28S rRNA bands. Then, 1 μg of each RNA sample was used as a template for cDNA synthesis using an iScript cDNA synthesis kit (1708891; Bio-Rad). Real-time polymerase chain reaction (PCR) was then performed using the iTaq Universal SYBR Green Supermix (1725124; Bio-Rad). Specific primers for each marker and for an endogenous control gene (18S rRNA) were designed using Gene Expression 1.1 software. Primer sequences were evaluated using Basic Local Alignment Search Tool (BLAST) to exclude those that would amplify undesired genes (). A universal Reference RNA consisting of 10 human cancer cell lines (740000-41; Agilent) [adenocarcinoma (mammary gland), hepatoblastoma (liver), adenocarcinoma (cervix), embryonal carcinoma (testis), glioblastoma (brain), melanoma, liposarcoma, histiocytic lymphoma (macrophage and histocyte), lymphoblastic leukemia (T lymphoblast), and plasmacytoma (myeloma and B lymphocyte)] as well as human genomic DNA (G304A; Promega) were used in quantitative PCR (qPCR) reactions. The PCR program was as follows: initial denaturation at 95°C, 50 cycles of denaturation at 95°C for 10 sec followed by annealing at 59°C for 30 sec. A final extension step was performed at 72°C for 10 min followed by melting curve analyses. Data were analyzed according to the method described in CitationLivak and Schmittgen (2001).
Knockdown
During the exponential phase of proliferation, colon CSCs were seeded in 24-well plates (E36112-39; Celprogen) and transfected with small interfering RNA (siRNA) specific for L1-ORF2 using Lipofectamine 2000 (11668-027; Invitrogen), according to the manufacturer's instructions. The siRNA was designed in accordance with the rules of CitationReynolds et al. (2004) and the sequence was as follows: L1-ORF2: 5’UAUUUCUGCCUUCAUUUCGdTdT 3’. The sequence was examined using BLAST to exclude sequences that would suppress undesired genes and ensure specificity. Following 48 h of incubation, cells were harvested by trypsinization (25200–072; Invitrogen). Samples incubated with Lipofectamine alone (without siRNA) were also tested to study the effect of compound alone on gene expression. Finally, samples incubated with non-specific siRNA were tested to study the specificity of the particular siRNA. The mRNA knockdown was calculated relative to a non-targeting control siRNA in each experiment. The expression level of the gene of interest and percentage knockdown was calculated using comparative Ct method:
Flow cytometry
Colon CSCs pre- and post-transfection were stained with phycoerythrin annexin V and 7-amino-actinomycin (559763; BD Biosciences) for 15 min and then resuspended in 0.5 ml of sheath fluid (8546859; Beckman Coulter) followed by flow cytometric analysis of more than 50,000 events. The data were analyzed using FCS Express Software (DeNovo).
Statistical analysis
The qPCR results were tested according to the Kolmogorov–Smirnov test; all samples had normal distribution. Median values were used for the analysis. Finally, Mann–Whitney tests were also performed on the qPCR data. All the reactions (molecular assays, knockdown, and flow cytometry) were performed in triplicates. A p value < 0.05 was considered significant.
RESULTS
Molecular assays
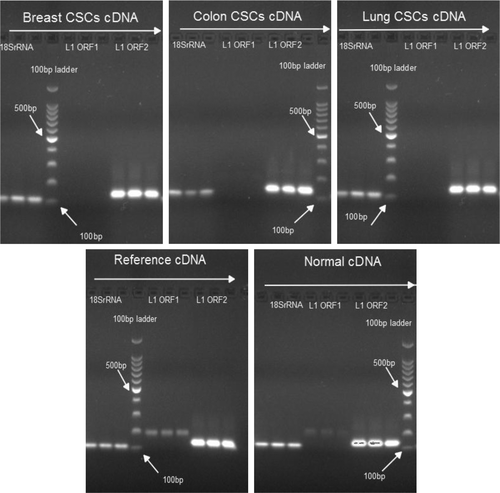
PCR analysis revealed that L1 ORF1 mRNA expressed in normal (non-cancerous) and cancer cell lines, but not in CSCs. In contrast, L1 ORF2 mRNA was present in all examined samples ().
Figure 1. Determining expression of L1 ORF1 and L1 ORF2. The presence of L1 ORF1 and L1 ORF2 mRNA was determined in CSCs, cancer cell lines, and non-cancerous (normal) samples. Expression of 18SrRNA was used as control. All reactions were performed in triplicates. The estimated length of PCR products is L1-ORF1: 187bp, L1-ORF2: 137bp, and 18srRNA: 112bp.
Of the EMT–MET markers that were studied, E-cadherin was expressed higher in cancer cells than in CSCs. N-cadherin gene expression levels were almost the same across all the tested cell lines. With the exception of lung cancer cell lines, vimentin expression levels were higher in CSCs, while β-catenin expression was higher than that of N-cadherin and vimentin in all cases. In normal (non-cancerous) samples, higher expression of vimentin was observed ().
Figure 2. Study of EMT–MET markers. The figure presents the qPCR data from CSCs, the respective cancer cell lines, reference cancer sample, and non-cancer samples. 18SrRNA served as a housekeeping gene in analysis. A lower ΔCt indicates higher gene expression.
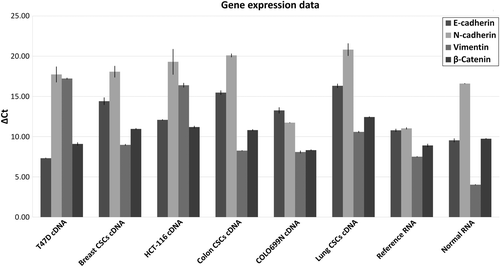
Various differences between gene expression data from cancer cells and CSCs were observed, with differences also noted when comparing different types of cancer. In breast and colon CSCs, SNAIL1, SNAIL2, zinc finger E-box-binding homeobox 1 (ZEB1), fribronectin 1 (FN1), and v-myc avian myelocytomatosis viral oncogene homolog (cMYC) were not expressed. In lung CSCs only ZEB1 was not expressed. For cancer cell lines, expression of N-cadherin was absent in HCT-116 cells, while in COLOO699N the SNAIL1 gene was not expressed.
siRNA transfection
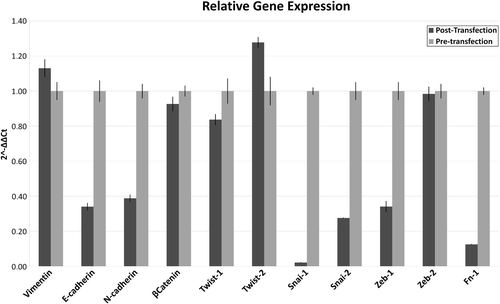
A knockdown percentage of up to 85% was achieved following treatment with L1-ORF2 siRNA. Compared with control cells, the decrease in gene expression was 2.78-fold for NANOG and 0.64-fold for SOX2 (). A decrease in expression was observed for almost all EMT–MET markers. The decrease was higher for SNAIL1, followed by SNAIL2, FN1, and E-cadherin. A small increase was observed for Vimentin and TWIST2 ().
Figure 3. Gene expression of stemness transcription factors. The figure presents the qPCR data before and after the knockdown of L1 ORF2. The control sample is the same cell line before siRNA transfection. Samples were normalized to control sample using Livak analysis, while 18SrRNA was used as housekeeping gene. The percentage knockdown of L1 ORF2 was up to 89%.
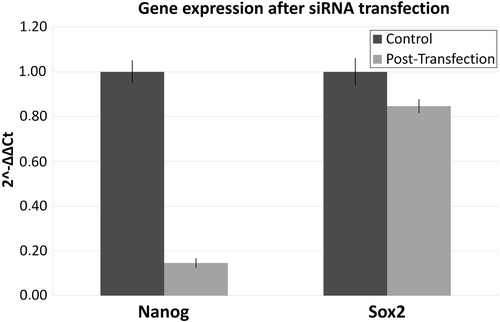
Figure 4. Gene expression of EMT–MET markers after knockdown of L1 ORF2. The figure presents the qPCR data before and after the knockdown of L1 ORF2. Samples were normalized to control sample using Livak analysis and 18SrRNA was used as housekeeping gene. The positive values represent increased gene expression, while a negative value decreased gene expression.
Cell cycle distribution
The number of cells undergoing apoptosis was almost 10 times higher following suppression of L1-ORF2 by siRNA. Under these same conditions, the number of dead cells observed was about 5-fold of that in control cells ().
Table 1. Cell cycle distribution.
Table 2. Primer sequences.
DISCUSSION
Cell plasticity is typically associated with the ability of stem cells to give rise to progenitor cells, which then mature into a variety of functional cell types. It occurs primarily during development, homeostasis, or as a response to injuries. However, plasticity may also occur when a differentiated cell type converts into another type of differentiated cell. Furthermore, progenitor or differentiated cells may be reprogrammed to more primitive cells (CitationTang, 2012).
Phenotypic plasticity in cancer cells is correlated with two important processes: EMT and MET. During EMT, epithelial cells lose cell-to-cell adhesion, migrate into the bloodstream, and become mesenchymal cells. This process is essential for mesoderm and neural tube formation, as well as other developmental processes. EMT is also involved in the initiation of metastasis and cancer progression. Primary tumor cells can invade into the bloodstream in a process mediated by E-cadherin. These CTCs move through the blood circulation and can regain an epithelial phenotype via MET and thus form micrometastases (CitationChaffer, 2011). According to literature and experimental data, CSCs can be formed by EMT induction. The expression of E-cadherin repressors in mammary epithelial cells in combination with TGF-β gives rise to cells with properties reminiscent of breast CSCs (CitationMani, 2008). Additionally, the EMT inducer ZEB1 represses microRNAs that inhibit CSCs maintenance (CitationWellner, 2009). Here, we confirm that cancer cells express high levels of E-cadherin and low levels of N-cadherin, explaining the epithelial phenotype of cancer cells. However, in contrast with other reports we found that E-cadherin gene expression was higher than that of N-cadherin. Nevertheless, vimentin gene expression was higher in CSCs than in cancer cells. These observations indicate that vimentin is essential for maintenance of the mesenchymal phenotype of CSCs, while E-cadherin is essential for maintenance of the epithelial phenotype of cancer cells.
CSCs initiate metastasis in breast cancer via EMT– MET processes (CitationTomaskovic, 2009). In breast CSCs, the TWIST1, TWIST2, and ZEB2 transcription factors are over-expressed, while in the T47D cancer cell line, expression of the transcription factors that repress E-cadherin is low. Furthermore, EMT is also involved in colorectal cancer and has an important role in tissue fibrosis (CitationSipos, 2012). The downregulation of E-cadherin in colon CSCs supports these observations and further supports a role for EMT in metastasis and cancer progression.
The role played by many signal transduction pathways in induction of EMT is now known (CitationRadisky, 2005). Consequently, targeting these pathways could reduce or impair the EMT process and thus reduce the incidence of micrometastases. Unfortunately, because of cross-talk between signaling pathways it is very difficult to suppress or induce a transcriptional factor located more downstream.
Therefore, a preferable target would not be involved in transduction pathways, but nevertheless still be capable of affecting the EMT process. Retrotransposons represent such a target, and approximately half (45%) of the human genome consists of retrotransposons.
LINE-1 retrotransposons comprise about 17% of human DNA (CitationLander, 2001). They have an internal polymerase II promoter and encode two ORFs. These transposons are responsible for most reverse transcription in the genome. These elements are actively transcribed and thus can influence chromatin remodeling events at the epigenetic level (CitationChueh, 2009). L1 retrotransposons cause genetic instability via chromosomal rearrangements and are able to induce apoptosis in breast cancer cells (Belgnaoui, 2012; CitationChen, 2012). A role for L1 in cancer is further supported by the observation that the L1 promoter is hypomethylated in many carcinomas (CitationTsutsumi, 2000).
Expression of LINE retrotransposons correlates with that of sex determining region Y (SRY) family factors, and it has been demonstrated that SRY-box (SOX) factors regulate the L1 transposable elements (CitationTchenio, 2000). Here we have demonstrated that L1 ORF2p gene expression is correlated with expression of the SOX2 transcription factor, which is involved in maintaining the stemness phenotype (CitationLeis, 2012). Suppression of L1 led to a decrease in SOX2 gene expression. However, we also observed a reduction in gene expression of another transcription factor essential for pluripotency, nanog homeobox (NANOG). The increase in gene expression of NANOG was higher than that for SOX2, confirming that the SOX2 and octamer-binding transcription factor 4 (OCT4) regulate the expression of NANOG (CitationRodda, 2005). Together, these data point to the role of the L1 retrotransposon in stemness regulation. According to CitationCarreira et al. (2014), the methylation of L1 promoter activates its transcription, which is followed by retrotransposition into an oncogenic region. After the tumorigenesis, the L1 promoter is hypomethylated, and this is correlated with an EMT that could lead to metastasis. In mouse, the L1 regulates nephrogenesis in vitro via both insertional and non-insertional mechanisms that disrupt MET (CitationRamos, 2011).
This study also identified a correlation between L1 expression and the EMT process. L1 ORF2p expression correlates with SNAIL1 and SNAIL2 genes expression, which in turn induce EMT. A reduction in L1 ORF2p expression led to a decrease in expression of the above genes in colon CSCs. The SNAIL1 and SNAIL2 can downregulate the E-cadherin and thus induce EMT (Battle, 2000). We further observed a reduction in N-cadherin and FN-1 genes; however, the gene expression levels of E-cadherin were not increased. Therefore, it is indicated that L1 interacts not only with SNAIL transcription factors, but also with E-cadherin. However, how they interact is not entirely understood.
CONCLUSION
The L1 retrotransposons are found in high copy numbers in the human genome and participate in genome rearrangement through transposition. ORF2p might be involved in maintaining stemness of colon CSCs through direct or indirect interaction with specific transcription factors essential for the process. ORF2p is also likely to be involved in EMT–MET by influencing gene expression of crucial EMT inducers, impacting upon the cellular plasticity of cancer cells. Analysis of L1 retrotransposon levels could be used either as a key tool in prognosis of cancer progression, or it could potentially serve as a novel druggable target. Further experiments should be performed in a range of cancer types in order to verify these concepts.
ACKNOWLEDGMENTS
The authors thank the two anonymous referees whose valuable commentary allowed us to improve the manuscript.
Declaration of interest: The authors report no declarations of interest. The authors alone are responsible for the content and writing of the paper.
References
- Kalluri R, Weinberg RA (2009). The basics of epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
- Chaffer CL, Weinberg RA (2011). A perspective on cancer cell metastasis. Science. 331:1559–1564.
- Lee JM, Dedhar S, Kalluri R, Thompson EW (2006). The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 172:973–981.
- Toloudi M, Apostolou P, Chatziioannou M, Papasotiriou I (2011). Correlation between Cancer Stem Cells and Circulating Tumor Cells and Their Value. Case Rep Oncol. 4:44–54.
- Lander ES, Linton LM, Birren B et al (2001). Initial sequencing and analysis of the human genome. Nature. 409:860–921.
- Feng Q, Moran JV, Kazazian HH, Jr., Boeke JD (1996). Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 87:905–916.
- Munz M, Baeuerle PA, Gires O (2009). The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 69:5627–5629.
- Chiou SH, Yu CC, Huang CY et al (2008). Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 14:4085–4095.
- Xiang R, Liao D, Cheng T et al (2011). Downregulation of transcription factor SOX2 in cancer stem cells suppresses growth and metastasis of lung cancer. Br J Cancer. 104:1410–1417.
- Janikova M, Skarda J, Dziechciarkova M et al (2010). Identification of CD133+/nestin+ putative cancer stem cells in non-small cell lung cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 154:321–326.
- Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A (2004). Rational siRNA design for RNA interference. Nat Biotechnol. 22:326–330.
- Tang SN, Fu J, Nall D, Rodova M, Shankar S, Srivastava RK (2012). Inhibition of sonic hedgehog pathway and pluripotency maintaining factors regulate human pancreatic cancer stem cell characteristics. Int J Cancer. 131:30–40.
- Mani SA, Guo W, Liao MJ et al (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715.
- Wellner U, Schubert J, Burk UC et al (2009). The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 11:1487–1495.
- Tomaskovic-Crook E, Thompson EW, Thiery JP (2009). Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res. 11:213.
- Sipos F, Galamb O (2012). Epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions in the colon. World J Gastroenterol. 18:601–608.
- Radisky DC (2005). Epithelial-mesenchymal transition. J Cell Sci. 118:4325–4326.
- Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH (2009). LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 5:e1000354.
- Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A (2006). Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 6:13.
- Chen L, Dahlstrom JE, Chandra A, Board P, Rangasamy D (2012). Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res Treat. 136:129–142
- Tsutsumi Y (2000). Hypomethylation of the retrotransposon LINE-1 in malignancy. Jpn J Clin Oncol. 30:289–290.
- Tchenio T, Casella JF, Heidmann T (2000). Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 28:411–415.
- Leis O, Eguiara A, Lopez-Arribillaga E, et al (2012). Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 31:1354–1365.
- Rodda DJ, Chew JL, Lim LH, et al (2005). Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 280:24731–24737.
- Carreira PE, Richardson SR, Faulkner GJ (2014). L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J. 281:63–73.
- Ramos KS, Montoya-Durango DE, Teneng I, Nanez A, Stribinskis V (2011). Epigenetic control of embryonic renal cell differentiation by L1 retrotransposon. Birth Defects Res A Clin Mol Teratol. 91:693–702.
- Batlle E, Sancho E, Franci C et al (2000). The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2:84–89.
