Abstract
The objective of this work was to investigate whether proteomic analysis of thymoma cells treated with the trichothecene deoxynivalenol (DON) as compared to non-treated (control) cells would reveal differential protein expression, and thus would contribute to a better understanding of the mechanisms of its toxicity. For that purpose the mouse thymoma cell line EL4 was exposed to 0.5 μM DON for 6 hr. A total of 30 proteins were affected after exposure of EL4 cells to DON. Most of these proteins were up-regulated and included key metabolic enzymes (e.g., fatty acid synthase, aldose reductase, carbamoyl phosphate synthetase, glucose-6-phosphate isomerase), chaperones (e.g., HSP9AB1 and HSP70), enzymes implicated in protein folding (PDI and ERO1-lα), and proteins involved in protein degradation (ubiquitin-conjugating enzyme (E1) and proteasome subunit alpha type-1). In addition, an IgE-binding protein with a molecular weight of 60 kDa and My-binding protein 1a (MYBBP1A), a transcription factor, were found to be up-regulated by DON. The observed up-regulation of MYBBP1A, a known repressor of a number of transcription factors such as PGC-1α, C-myb, and p65 of the NF-κB family, suggests that this protein might play a role in the mechanism of DON toxicity.
Introduction
Trichothecenes represent a family of toxins of fungal origin, also referred to as mycotoxins (Rocha et al., Citation2005). The trichothecene deoxynivalenol (DON) is produced by Fusarium species and is detected in fungal contaminated corn, wheat, barley, and other cereals worldwide which are intended for livestock and human consumption (Rocha et al., Citation2005). DON, known also as vomitoxin, is of health concern both for human beings and animals (Mikami et al., Citation2004; Rocha et al., Citation2005; Chidozie et al., Citation2008). Extensive studies of this toxin have been conducted in vivo and in vitro (Tomar et al., Citation1986; Minervini et al., Citation2004; Pinton et al., Citation2008). The immune system is considered a prime target of trichothecene toxicity (Pestka and Bondy, Citation1994; Pestka, Citation2003) and, depending on the dose and exposure frequency, can be either suppressive or stimulatory (Yang et al., Citation2000).
Regarding the mechanism of toxicity, several studies have shown that DON binds to ribosomal peptidyl transferase of the 28S rRNA, thus inhibiting protein synthesis (Shifrin and Anderson, Citation1999). Indeed, a number of toxicants, including a group of trichothecenes (like DON), some antibiotics (like anisomycin), and palytoxin (a non-peptide marine toxin), were found to induce a phenomenon described as ribotoxic stress response (Iordanov et al., Citation1997); however, not all of these agents bind to the peptidyl transferase (Iordanov et al., Citation1997, Citation1998; Iordanov and Magun, Citation1998; Shifrin and Anderson, Citation1999). Nevertheless, in most cases, experimental evidence indicates that active ribosomes are required to transduce the stress signal (Iordanov et al., Citation1997). This stress signal can trigger the activation of mitogen-activated protein kinases (MAPKs), like JNK (c-Jun N-terminal kinases, a family of kinases that are activated by cell stress-inducing stimuli), which may lead to anti-proliferation and even result in apoptosis (Shifrin and Anderson, Citation1999; Moon and Pestka, Citation2002; Zhou et al., Citation2005). It is not yet known, however, how the signal transduces from the ribosomal site to the components of the signaling cascade that mediate the stress response (Zhou et al., Citation2005). Furthermore, it has also been shown that DON impairs mitochondrial oxidative metabolism in cell lines (Minervini et al., Citation2004) again, the mechanisms underlying this cytotoxic effect remain undefined.
The aim of the present study was to get a better insight into the mechanism of DON toxicity following a proteomics approach. Because EL4 cells have been shown to possess an immature thymus cell phenotype (Tanaka et al., Citation1987), they are used as a model system for thymocytes in mechanistic studies (Li et al., Citation2000; El-Darahali et al., Citation2005; Lee et al., Citation2008). The protein expression profile of EL4 cells exposed to DON was analyzed and examined for whether the exposure would result in differential protein expression (as compared to the untreated cells). Proteins extracted from treated and control cells were separated by one-dimensional (1D)-SDS-PAGE. Proteins bands were extracted, digested with trypsin, and the resulting peptide mixtures analyzed by nanoflow liquid chromatography coupled to a mass spectrometer. The relative abundance of the identified proteins was estimated using observed peptide hits. This quantitative approach was successfully employed earlier for biomarker discovery in urine (Pang et al., Citation2002), for estimating changes in protein expression levels in yeast in function of carbon source (Gao et al., Citation2003) and, recently, we have applied it in evaluating protein expression changes of thymoma EL4 cells exposed to bis-(tri-n-butytin)oxide (TBTO) (Osman et al., Citation2009). In the present study, the expression levels of 30 proteins were affected. The outcome of this study will be discussed in the context of already-reported data dealing with the mechanism of DON toxicity.
Methods and materials
Chemicals
Deoxynivalenol, DL-dithiothreitol (DTT), and iodoacetamide were purchased from Sigma (Mannheim, Germany). Ammonium bicarbonate was obtained from Fluka (Zwijndrecht, the Netherlands). ReproSil-PurC18-AQ3 was supplied by Dr. Maisch GmbH (Ammerbuch, Germany) and C18 200A-AQ5 was from Phenomenex (Uetrecht, the Netherlands). Solvents ethanol (absolute) acetonitrile (ACN, HPLC Grade) were purchased from Bissolve (Amsterdam, the Netherlands). Sequencing-grade modified trypsin was purchased from Roche (Mannheim, Germany) and the gel for electrophoresis (10% Nu-PAGE Bis–Tris-gel) was obtained from Invitrogen Company (Breda, the Netherlands).
Cell culture and exposure
Thymoma EL4 cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) and contained 1% penicillin (10,000 U/mL) and streptomycin (10,000 μg/ mL) (Fisher, Landmeer, the Netherlands). The total number of cells used per sample was 2.7 × 107 cells, suspended in 40 mL medium. The experiments for both treated and control samples were performed in duplicate. A stock solution of DON was prepared in ethanol (1 mM). For the exposures, 20 μL of this stock solution was added to 40 mL cell suspension of each treated sample so that the final concentration of DON was 0.5 μM, whereas 20 μL of ethanol without the mycotoxin was added to each control. The treated and the control samples were then incubated at 37°C for 6 hr. At a 0.5 μM concentration over a 6-hr period, the viability of the treated samples remained comparable to that of the untreated controls. Cell viability was evaluated by Trypan blue dye and was found to be above 80% in all experiments.
Protein extraction
The cell suspensions were centrifuged at 1200 rpm for 10 min at 4°C. The cell pellets were suspended in 20 mL of phosphate-buffered saline (PBS, pH 7.2) and centrifuged. This washing step was repeated once more before each of the cell pellets (treated and control) was suspended in 2 mL of 100 mM Tris–HCl (pH 8) containing 5 mM EDTA and 1% SDS (sodium dodecyl sulphate). The samples were then sonicated in 4 cycles of 10 sec; each 5-sec sonication was followed by a 5-sec break. Subsequently, the samples were centrifuged at 11,000 rpm for 20 min at 4°C. The supernatants obtained were recovered and kept at −20°C until use. Aliquots of the supernatants (before freezing) had their protein concentration evaluated using the Bradford method (Bradford, Citation1976). The protein concentrations of the treated samples and the controls were found to be 1.75 and 1.9 mg/mL (average value of duplicates), respectively.
Gel-electrophoresis
One-dimensional-SDS-PAGE gel was used for the separation of proteins. Eighty micrograms of the exposed sample and the control were loaded onto wells of a 10% Nu-PAGE-Tris-gel. After the gel had been run at a constant voltage of 160 V for 90 min, it was stained of using Coomassie stain (SimplyBlue Safe Stain), according to the instructions of the Invitrogen Company. The gel was rinsed three times with 100 mL of deionized water for 5 min to remove SDS and salts. This was followed by staining the gel with 20 mL of the Coomassie stain for 2 hr. The gel was kept in deionized water (at maximum) overnight. After that, the lanes of the sample and the control were cut horizontally into 15 slices. Each slice was excised from the gel with a razor blade, cut into small pieces, and then transferred into 1.5 mL Eppendorf tubes.
In-gel digestion
The fractions were de-stained with 50 mM NH4HCO3/50% ethanol solution for 25 min. This procedure was performed twice, followed by dehydration of the gel with 100% ethanol. Proteins were reduced with 10 mM DTT (37°C, 45 min) and then alkylated with 55 mM iodoacetamide at 37°C in the dark for 45 min. This was followed by dehydration of the gel pieces with 100% ethanol and air-drying for 15 min. Subsequently, the gel was covered with 30 μL of modified trypsin solution (12.5 ng/μL) in 50 mM ammonium bicarbonate buffer. After incubation for 17 hr, an additional 30 μL of buffer was added to each fraction and incubation continued for a further for 2 hr. The supernatant of each fraction was collected into a clean Eppendorf tube and 25 μL of 50% ethanol containing 5% formic acid was added to each fraction, mixed well and pooled it to the previous one. This latter step was repeated once. The fractions were then dried in a SpeedVac, suspended in 60 μL of 100% acetonitrile and dried again in a Speed Vac. Last, each fraction was dissolved in 20 μL of 2% ACN/10% formic acid solution. The samples were kept at −20°C until use for nano-LC/MS/MS analysis.
Nano-LC/MS/MS analysis
Nano-LC/MS/MS analysis was performed by using a system consisting of an Agilent 1100 Series LC set up (vacuum degasser, autosampler and one high pressure-mixer binary pump without static mixer) coupled to a LCQ Deca Qaudruple Ion Trap mass spectrometer (Thermo Finnigan, San Jose, CA) as described earlier (Meiring et al., Citation2002). Briefly, 5 μL peptide solution was delivered to a trap column [Aqua C18 200A (Phenomenex); l = 15 mm, 100 μm ID, packed in-house] at 5 μL/min of 100% Solvent A (0.1 M acetic acid). After decreasing the flow to ≈150 nL/min by a splitter, the peptides were transferred to the analytical column [Reprosil C18RP (Dr.Maisch GmbH); l = 20 cm, 50 μm ID packed in-house] with a linear gradient from 0% to 100% Solvent B (0.1 M acetic acid in 80% ACN) in 40 min. The 100% B was maintained for 5 min, followed by decreasing Solvent B to 0% in 0.1 min, washing and re-equilibrating the system at 100% A for 10 min. The column eluent was sprayed directly into the ESI source of the mass spectrometer via a butt-connected nano-ESI emitter (New Objectives, Inc., Woburn, MA). The LCQ operated in a positive ion mode and the peptides were fragmented in a data-dependent mode. A full MS scan was followed by tandem spectra of the three most intense precursor ions present in the MS scan. The capillary voltage was 1.9 kV and capillary temperature was 200°C.
Database searches
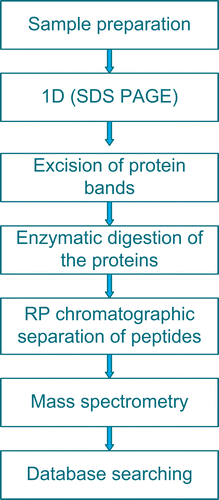
All MS/MS data was searched using SEQUEST engine (Bioworks version 3.3) (Yates et al., Citation1995) against the IPI mouse v 3.18 fasta database. The searches were performed allowing for fixed modification on methionine (oxidation) and cysteine (carbamidomethylation) residues. The peptide and fragment mass tolerance were set at 1.4 and 1 Da, respectively. Trypsin was used for protein digestion and two missed cleavages were allowed. For protein identification acceptance, the following filtering criteria were used: a Delta Cn (δCn) was set to 0.1 SEQUEST Xcorr versus charge state (Xcorr = 2 for z = 1, Xcorr = 1.5 for z = 2 and 3.3 for z = 3 and a peptide probability score <0.01. The Xcorr score is a measure of how well the experimental mass spectrum matches the spectrum generated in silico. δCn measures how different the first identification is from the next best. Protein identification was also based on at least two different peptides. The total identified proteins were obtained by combining the identified proteins of both the samples and the controls. The following criteria were applied to determine whether or not DON affected the expression level of a protein in the EL4 cells: A 1.4-fold change or greater in the expression level of a protein and consistency in the direction of change in both experiments were required. Further, if a protein was not detected, e.g., in an exposed sample, while it is present in the corresponding control, or vice versa, then a value of one peptide was assumed for that protein. The workflow of a typical proteomic experiment of the present study is summarized in .
Results
Separation of proteins by one-dimensional electrophoresis analysis
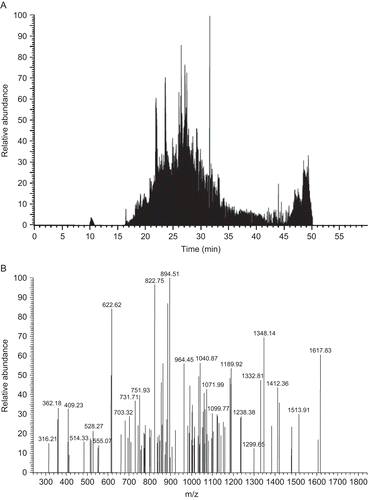
Upon separation of proteins obtained from DON-treated and the control cells by 1D-SDS-PAGE, the protein bands were digested with trypsin and the resulting peptide mixtures were analyzed by a nano-LC/MS/MS. , for instance, illustrates the total ion chromatogram of tryptic peptides obtained from fraction 8 of DON-treated extract in Experiment I, whereas illustrates tandem MS/MS of the peak at retention time 26.04 min.
Protein identification
The criteria used for identification of the proteins were described in the section “Methods and Materials.” Because the number of proteins identified in the controls was less compared to those of the exposed samples, analysis of each control was repeated once (see on-line Supplementary Table). This re-analysis of the control samples resulted in identification of proteins that were not detected in the first runs. These recovered proteins were taken into account in the data analysis. As reported in , 524 proteins were identified in Experiment I. This total number of proteins was obtained by combining the proteins identified both in the treated sample and corresponding control. About 61% of these identified proteins (319) were present in both the treated sample and its control (); ≈22% (116) of the proteins were found in the control and ≈17% (89) of the proteins were found in the DON-treated sample of Experiment I ().
Figure 3. (A) Illustrates the number of proteins identified in both the DON-exposed sample and the control of Experiment I, as well as the proteins detected only either in the DON-treated sample or in the control. To obtain the total proteins identified in Experiment I, proteins present in the control and in the sample were combined. (B) Indicates the number of proteins identified that are common to both sample and control of Experiment II, and the proteins detected either in the sample or in the control. The total proteins identified in Experiment II were obtained by combining proteins present in the sample and in the control.
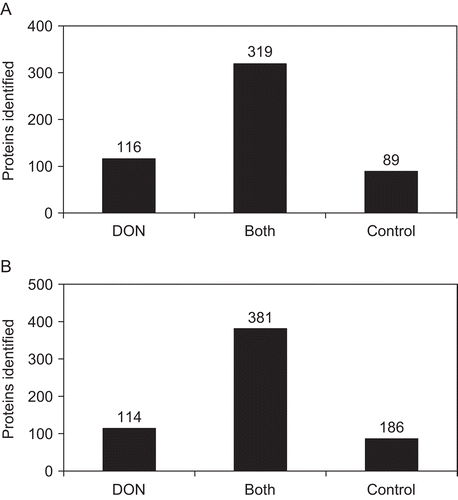
In Experiment II, the total number of proteins identified was 681 (); again, that number resulted from the combination of proteins found in both the treated sample and its corresponding control. Circa 56% (381) were present in both treated sample and control. Twenty eight percent (186) were found in the control and 17% in the treated sample of Experiment II (). The combination of all the proteins identified in both experiments (treated samples and controls) led to 790 proteins (see on-line Supplementary Table).
To evaluate the effect of DON on the expression levels of proteins identified in EL4 cells, we used a semi-quantitative proteomics method based on the number of peptide hits obtained, which reflects the relative protein abundance (Rappsilber et al., Citation2002). Therefore, for a given protein, we determined the ratio of the peptides identified for that protein in the treated sample and in the corresponding control.
Proteins affected by the exposure of EL4 cell line to DON
We set two criteria to attribute a specific affect on a protein’s expression to DON exposure (see section “Materials and Methods”). A total of 30 proteins fulfilled the criteria set and were therefore considered specifically altered by treatment of EL4 cells with DON ( and ). Nine of these proteins were not detectable in control cells (). The differentially-expressed proteins can be divided into six groups according to their functions: (1) metabolic enzymes; (2) chaperones and enzymes involved in protein folding; (3) protein synthesis, mRNA processing proteins and a cyto-linker; (4) ubiquitin-proteasome system and proteins implicated in immune response; (5) proteins involved in cell trafficking; and, (6) proteins implicated in transcription and DNA replication. It should be noted that one of the up-regulated proteins (1.5-fold, mean value), Isoform 2 of protein C1orf77 homolog (), was not placed in either of the six groups as its function is unknown. The sequence of this protein was previously deduced from mouse brain cDNA clones (Chambers and Abbot, Citation1996) and corresponds to hypothetical protein LOC66511 (with gene symbol 2500003M10 Rik as indicated by a BLAST database search).
Table 1. Identified proteins affected by the exposure of EL4 cells to deoxynivalenol (DON).a
Table 2. List of the proteins detected only in DON-exposed cells.
Metabolic enzymes
Expression levels of enzymes such as aldose reductase, fatty acid synthetase, carbamoyl phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (a multi-functional enzyme), ATP synthase-β chain, mitochondrial precursor, and glucose-6-phosphate isomerase were up-regulated in the DON-treated cells (). Aldose reductase was up-regulated 1.9-fold, whereas fatty acid synthetase (major enzyme involved in lipid metabolism) was increased 4.8-fold. Carbamoyl phosphate synthatase, aspartate ttranscarbamylase, and dihydroorotase, which constitute a multifunctional (consists of three activities) enzyme also known as CAD, catalyze the first three reactions of the de novo synthesis of pyrimidine nucleotides (Huang and Graves, Citation2003); this complex protein was increased by 2.4-fold. The expression levels of both β-subunit of the mitochondrial H+-ATP synthetase and the phosphoglucose isomerase (GPI) were up-regulated 2.9-fold and 1.5-fold, respectively. The former activity catalyzes the rate-limiting step of ATP formation in eukaryotic cells (Izquierdo, Citation2006) and the latter accelerates the second step of the glycolysis pathway (Tsutsumi et al., Citation2003). Further, GPI also acts as a cytokine, a function that is independent of its enzymatic role (Tsutsumi et al., Citation2003).
Table 3. List of the up- and down-regulated proteins with gene symbols, peptide ratios* and the pathways implicated.
Chaperones and enzymes involved in protein folding
It was also found here that the expression levels of chaperones and enzymes involved in protein folding processes were altered in the DON-exposed cells. Chaperones HSPA9 (stress-70 protein, mitochondrial precursor), HSP9AB1 (heat shock protein 1, 84 kDa), T-complex protein 1 subunit-theta, and PRDX2 (peroxiredoxin-II) were increased by 2.0-, 2.3-, 4.0-, and 2.6-fold, respectively ( and ). In contrast, heat shock protein 1A (HSPA1A) was decreased by 2.0-fold (). The HSPs are known to protect intracellular proteins so that proteins maintain their normal conformations in the presence of stress-inducing agents (Gething and Sambrook, Citation1992), whereas peroxiredoxin II is a member of ubiquitous thiol-specific antioxidant enzymes that protect cells not only from peroxides, but are also implicated in other cellular functions such as protection of radical-sensitive proteins, and intracellular signal transduction (Lee et al., Citation2003) and the references therein]. Further, enzymes implicated in protein folding pathway in the endoplasmic reticulum exhibited higher expression levels in the DON-treated cells. Protein-disulfide isomerase family A, member 3 (PDIA3) and Protein-disulfide isomerase family A, member 6 (PDIA6) were both up-regulated by 1.6-fold, whereas Endoplasmic oxido-reductin-1 (ERO1-α) was increased by 1.8-fold (). PDIs catalyze thiol disulfide interchange reactions (Laboissière et al., Citation1995; Mezghrani et al., Citation2001), whereas the ERO1-α catalyzes the regeneration of the reduced form of the protein-disulfide isomerase (Mezghrani et al., Citation2001).
Membrane transport proteins
Among the proteins detected in the DON-treated cells but not in the controls were two proteins implicated in vesicle-mediated cellular transport: isoform 1of Septin-9 and Rab GDP-dissociation inhibitor-β (). The former protein belongs to the septin family that exhibit multiple cellular activities, including cytokinesis (Field and Kellog, Citation1996), membrane transport (Beites et al., Citation1999), and control of cell polarity (Drees et al., Citation2001). GDP-dissociation inhibitor-β modulates translocation of Rab proteins between cytosol and cell membranes (Wu et al., Citation2007), and thereforeplays an important role in regulation of intracellular vesicle traffic.
Protein synthesis, mRNA processing proteins and a cyto-linker
Proteins involved in mRNA processing and in protein synthesis were also up-regulated. These included ribosomal proteins (1.9–4.0-fold), heterogeneous nuclear ribonucleoprotein A/B (1.7-fold) and eukaryotic translation initiation factor 3 subunit 1 (2.0-fold) (). Moreover, the expression level of plectin 8, a cyto-linker was increased 3.0-fold (). This cyto-linker protein has not only structural role in the cell, but also may play a dynamic role in the transduction of cellular signals (Osmanagic-Myers and Wiche, Citation2004).
Ubiquitin-proteasome system and immune response-related proteins
DON also affected proteins involved in protein degradation pathway. The expression level of ubiquitin-conjugating enzyme 1 (UBELY1) was increased in DON-treated cells (2-fold). Likewise, alpha subunit 1 of the proteasome complex (PMSA1) was also up-regulated 2.2-fold (). The ubiquitin-proteasome system plays an important role in the turnover of damaged proteins (Chen and Madura, Citation2005), but also regulates growth and stress responses (Chen and Madura, Citation2005). Interestingly, we also identified an IgE-binding protein with a molecular weight of 60 kDa in the thymoma EL4 cell line. This protein was up-regulated 1.8-fold in the DON-treated cells ().
Proteins involved in transcription and DNA replication
Other proteins whose expression levels were up-regulated by DON are: the mini-chromosome maintenance 6 protein (MCM6; important protein in proliferation and cell cycle) (Barton and Levine, 2008) and histone 2B, an integral part of chromatin (). Both proteins were not detectable in the control cells. Finally, two transcription factors, RBMX and Myb-binding protein 1A (MYBBP1A), were affected by DON. Expression levels of these proteins were increased by 2.0- and 2.4-fold, respectively in the DON-exposed EL4 cells ().
It has been suggested that in mouse liver RBMX is a transcriptional regulator of sterol regulatory element-binding protein-1c (SREBP1c) gene expression (Takemoto et al., Citation2007). MYBBP1A is a protein predominantly localized in nucleoli (Tavner et al., Citation1998) and has recently been shown to be a repressor of a number of transcription factors (Jones et al., Citation2002; Fan et al., Citation2004; Owen et al., Citation2007). This latter protein, whose processing and translocation have both been shown to be induced upon ribosome stress (Yamauchi et al., Citation2008), may play an important role in DON toxicity.
Discussion
In the present study, we performed proteomic analysis of thymoma EL4 cells exposed to one of major toxins of fungal origin, DON. We found that DON affected expression levels of 30 proteins ( and ); most of these were up-regulated ( and ). This is in agreement with previous reports wherein DON was shown mainly to induce genes and down-regulate only a few (Nishiuchi et al., Citation2006; Boddu et al., Citation2007). The proteins whose expression levels were increased in the DON-treated cells include enzymes that catalyze key metabolic pathways. One affected enzyme is aldose reductase, the first enzyme in the polyol pathway, which several previous studies have shown to be induced in osmotic related stress (Galvez et al., Citation2003). DON is known to affect cell membrane permeability (Kasuga et al., Citation1998), and the mycotoxin T-2 (another trichothecene) is known to alter cell volume and shape (Holt et al., Citation1988).
The expression level of fatty acid synthetase, another key enzyme, was increased in the DON-exposed cells (). A possible explanation for this may be that DON produces hydrogen peroxide, a known inducer of the enzyme (Furuta et al., Citation2008). A number of studies showed that DON causes lipid peroxidation and consequently produces reactive oxygen species, including hydrogen peroxide (Desmond et al. Citation2008, and references therein). Alternatively, the observed up-regulation of fatty acid synthetase might be mediated by RBMX, one of the up-regulated proteins identified here ( and ). In fact, this RBMX has been previously shown to bind the SREBP-1c promoter, regulating positively expression of this gene (Takemoto et al., Citation2007). SREBP-1 is known to be involved in up-regulation of fatty acid synthetase (Furuta et al., Citation2008).
Among other enzymes up-regulated in the DON-treated cells were GPI, the β-subunit of mitochondrial ATP synthetase, and CAD (). Increased levels of these enzymes reflect the need to meet the energy and pyrimidine demand for the noted DON-induced protein synthesis. This would be in accord with the increased expression level of mini-chromosome maintenance (MCM6) protein, which reflects an increased proliferating state of the exposed cells (Barton and Levine, Citation2008). It would also be in agreement with the observed increased level of the eukaryotic translation initiation factor-3 () that plays a central role in the formation of 40S initiation complex and is associated with increased rates of protein synthesis (Miyamoto et al., Citation2005). However, GPI may also play a non-enzymatic role (it is known that lectin-stimulated T-cells release this protein) and is implicated in the production of immunogloblins in B-cells (Chaput et al., Citation1988). That DON induces the formation of immunoglobulins both in vivo and in vitro was reported before (Pestka et al., Citation2004, and references therein). We also identified an IgE-binding protein that is identical in sequence to a protein previously detected in 23B6 (a rat-mouse T-hybridoma cell line) culture supernatants and was shown to potentiate IgE responses (Martens et al., Citation1985). This IgE-binding protein might mediate the previously-reported DON stimulation of IgE production (Pestka et al., Citation2004).
Furthermore, chaperones (HSPA9, HSP90AB1) and protein folding enzymes (PDI and ERO1-lα) exhibited increased expression levels in DON-exposed EL4 cells compared to in non-treated cells (). This increase in the expression levels of these proteins might be the response to increased amount of cargo proteins destined for the secretory pathway in the endoplasmic reticulum (ER), following the increase of DON-induced protein expression levels. This can be related to the observed increased expression of Rab GDP-dissociation inhibitor-β and septin-9, proteins that play an essential role in protein transport (Beites et al., Citation1999; Wu et al., Citation2007) in the exposed cells. As shown in , protein disulfide isomerase oxidizes the SH groups of the proteins to disulfide bond; the reduced form PDI is then re-oxidized by ERO1-α. We cannot, however, rule out that the increased levels of chaperones might be due to DON-induced oxidative stress. Oxidative stress is known to be one of the principal mechanisms that lead to protein mis-folding (Chondrogianni et al., Citation2005).
Figure 4. A scheme illustrating the oxidative protein folding pathway in the endoplasmic reticulum (ER) (adapted from Tu and Weissman, Citation2004). DON induced the oxido-reductants, protein disulfide isomerase (PDI), and endoplasmic oxido-reductin-1 (ERO1-l) which catalyze this pathway. An increase in protein cargo (synthesis) or increase in mis-folded proteins (e.g., under oxidative stress) stimulates this pathway.

In addition, DON increased expression levels of some components of the ubiquitin-proteasome pathway (). This system consists of ubiquitin, a 76-amino acid polypeptide, three conjugating enzymes known as ubiquitin-conjugating (E1), (E2), and (E3), and a multiunit complex protease (i.e., the proteasome) (Naujokat and Hoffman, Citation2002; Chen and Madura, Citation2005). This is a tightly- and highly-regulated protein degradation pathway in which both normal and damaged proteins are degraded. One of the proteasome α subunits (PSMA1) and ubiquitin-conjugating E1 were up-regulated in the DON-treated cells (). Over-expression of these proteins in cells under stress conditions were noted before (Chen and Madura, Citation2005). Further evidence supporting our result comes from a recent study in which DON-treatment was linked to induction of genes encoding proteins involved in ubiquitination (Boddu et al., Citation2007).
Finally, a Myb-binding protein 1a (MYBBP1A) was identified that might be involved in DON toxicity. This transcription factor [MW = 160 kDa; predominantly localized in nucleolus (Tavner et al., Citation1998)], acts as a repressor of several transcription factors and activator for others (Jones et al., Citation2002; Fan et al., Citation2004; Owen et al., Citation2007). In some cells, MYBBP1A is processed into two 67 and 140 kDa products. The treatment of HeLa S3 cell lines with actinomycin D or with UV led to processing of MYBBP1A and its translocation from the nucleolus to nucleoplasma (Yamauchi et al., Citation2008); these events are induced upon ribosome stress. Products formed from MYBBP1A act as repressors of transcription factor PGC-1α, critical for mitochondrial biogenesis and function (Yamauchi et al., Citation2008). Because it has been shown that MYBBP1A represses PGC-α, a positive regulator of mitochondrial oxidative metabolism (Tavner et al., Citation1998; Lin et al., Citation2005), and because DON impairs mitochondrial activity (Minervini et al., Citation2004), it is likely that MYBBP1A contributes to the mechanism of DON toxicity.
Furthermore, since DON has been shown to activate mitogen-activated protein kinase (MAPKs) (Shifrin and Anderson, Citation1999; Moon and Pestka, Citation2002; Zhou et al., Citation2005), it is probable this activation occurs via MYBBP1A, as the latter has been shown to bind to c-jun (Yamauchi et al., Citation2008), thereby forming a bridge between the ribosomal site and effectors of the signaling cascade that mediate the stress response. This is consistent with the observed DON-induced phosphorylation of c-jun (Pestka et al., Citation2004) that subsequently triggers downstream signals that lead to the described immunotoxic effects of the trichothecene. Taken together, this study has shown that DON stimulated the expression levels of several proteins in thymoma cells. These proteins included key metabolic enzymes, proteins associated with the ubiquitin-proteasome system, immune-associated proteins, as well as a known transcription factor (MYBBP1A), each of which could play a role in the trichothecene mechanism(s) of toxicity.
Declaration of interest
This work has been funded by the Netherlands Genomics Initiative Organization for Scientific Research (NOW): nr 050-060-510.
Supplementary Material
Download PDF (1.3 MB)References
- Barton, K. M., and Levine, E. M. 2008. Expression patterns and cell cycle profiles of PCNA, MCM6, Cyclin D1, Cyclin A2, Cyclin B1, and phosphorylated histone H3 in the developing mouse retina. Devel. Dyn. 237:672–682.
- Beites, C. L., Xie, H., Bowser, R., and Trimble, W. S. 1999. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nature Neurosci. 2:434–439.
- Boddu, J., Cho, S., and Muehlbauer, G.J. 2007. Transcriptome analysis of trichothecene-induced gene expression in barley. Mol. Plant-Microbe Interact. 20:1364–1375.
- Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein dye binding. Anal. Biochem. 72:248–254.
- Chambers, D. M., and Abbot, C. M. 1996. Isolation and mapping of novel mouse brain cDNA clones containing trinucleotide repeats and demonstration of novel alleles in recombinant inbred strains. Genome Res. 6:715–723.
- Chaput, M., Claes, V., Portelle, D., Cludts, Cravador, A., Burny, A., Gras, H., and Tartar, A. 1988. The neurotrophic factor neuroleukin is 90% homologous with phosphohexose isomerase. Nature 332:454–455.
- Chen, L., and Madura, K. 2005. Increased proteasome activity, ubiquitin-conjugating enzymes, and eEFlA translation factor detected in breast cancer. Cancer Res. 65:5599–5606.
- Chidozie, J.A., Harkema, J.R., and Pestka, J.J. 2008. Tissue distribution and pro-inflammatory cytokine induction by the trichothecene deoxynivalenol in the mouse: Comparison of nasal vs. oral exposure. Toxicoloy 248:39–44.
- Chondrogianni, N., Tzavelas, C., Pemberton, A. J., Nezis, I. P., Rivett, A. J., and Gonos, E. S. 2005. Over-expression of proteasome β5 subunit increases the amount of assembled proteasome and confers ameliorated response to oxidative stress and higher survival rate. J. Biol. Chem. 280:11840–11850.
- Desmond, O.J., Manners, J.M., Stephens, A.E., Maclean, D.J., Schenk,P. M., Gardiner, D. M., Munn, A. L., andKazan, A. K. 2008. The Fusarium mycotoxin deoxynivalenol elicits hydrogen peroxide production, programmed cell death and defense response in wheat. Mol. Plant Pathol. 9:435–445.
- Drees, BL., Sundin,B., Brazeau, E., Caviston, J. P., Chen, G. C.,Guo, W., Kozminski, K. G., Lau, M. W., Moskow, J. J., Tong, A., Schenkman, L. R., McKenzie, A., 3rd,Brennwald, P., Longtine, M., Bi, E., Chan, C., Novick, P., Boone, C., Pringle, J. R., Davis, T. N., Fields, S., andDrubin, D. G. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154:549–571.
- El-Darahali, A., Fawcett, H., Mader, J. S., Conrad, D. M., and Hoskin, D. W. 2005. Adenosine-induced apoptosis in EL-4 thymoma cells is caspase-independent and mediated through a non-classical adenosine receptor. Exp. Mol. Pathol. 79:249–258.
- Fan, M.,Rhee, J., St-Pierre, J., Handschin, C., Puigserver, P., Lin, J., Jäeger, S., Erdjument-Bromage, H., Tempst, P., and Spiegelman, B.M. 2004. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: Modulation by p38 MAPK. Genes Dev. 18:278–289.
- Field, CM, and Kellog, D. 1996. Septins: Cytoskeletal polymers or signaling GTPases? Rev. Trends Cell Biol. 10:387–394.
- Furuta, E., Pai, S. K., Zhan, R., Bandyopadhyay, S., Watabe, M., Mo, Y. Y, Hirota, S., Hosobe, S., Tsukada, T., Miura, K., and Watabe, K. 2008. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 68:1003–1011.
- Galvez, A. S., Ulloa, J. A., Chiong, M., Criollo, A., Eisner, V., Barros, L.F., and Lavandero, S. 2003. Aldose reductase induced by hyperosmotic stress mediates cardiomyocyte apoptosis: Differential effects of sorbitol and mannitol. J. Biol.Chem. 278:38484–38494.
- Gao, J. I., Opiteck, G. J., Friedrichs, M. S., Dongre, A. R., and Hefta, S. A. 2003. Changes in the protein expression of yeast as a function of carbon source. J. Proteome Res. 2:643–649.
- Gething, M. J., and Sambrook, J. 1992. Protein folding in the cell. Nature 355:33–45.
- Holt, P. S., Buckley, S., Norman, J. O., and Deloach, J. R. 1988. Cytotoxic effect of mycotoxin on cell lines in culture as determined by a rapid colorimetric bioassay. Toxicon 26:549–558.
- Huang, M., and Graves, L. M. 2003. De novo synthesis of pyrimidine nucleotides: Emerging interfaces with signal transduction pathways. Cell. Mol. Life Sci. 60:321–336.
- Iordanov, M. S., and Magun, B. E. 1998. Loss of cellular K+ mimics ribotoxic stress inhibition of protein synthesis and activation of the stress kinase SEK1/MKK4, stress-activated protein kinase 1, and p38/HOG1 by palytoxin. J. Biol. Chem. 273:3528–33534.
- Iordanov, M. S., Pribnow, D., Magun, J. L., Dinh, T. H., Pearson, J. A., and Magun, B. E. 1998. Ultraviolet radiation triggers the ribotoxic stress response in mammalian cells. J. Biol. Chem. 273:15794–15803.
- Iordanov, M. S., Pribnow, D., Magun, J. L., Dinh,T. H., Pearson, J. A., Chen, S. L., and Magun,B. E. 1997. Ribotoxic stress response: Activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the α-sarcin/ricin loop in the 28S r RNA. Mol. Cell. Biol. 17:3373–3381.
- Izquierdo, J. M. 2006. Control of the ATP synthase β subunit expression by RNA-binding proteins TIA-1, TIAR and HuR. Biochem. Biophys. Commun. 348:703–711.
- Jones, L. C., Okino, S. T., Gonda, T. J., and Whitlock, J. P. Jr., 2002. Myb-binding protein 1a augments AhR-dependent gene expression. J. Biol. Chem. 277:22515–22519.
- Kasuga, F., Hara-Kudo, Y., Saito, N., Kumagai, S., and Sugita-Konishi, Y. 1998. In vitro effect of deoxynivalenol on the differentiation of human colonic cell lines Caco-2 and T84. Mycopathologia 142:161–167.
- Laboissière, M. C., Sturley, S. L., and Raines, R. T. 1995. The essential function of protein-disulfide isomerase is to unscramble non-native disulfide bonds. J. Biol. Chem. 270:28006–28009.
- Lee, T. H., Kim, S. U., Yu, S. L., Kim, S. H., Park, D. S., Moon, H. B., Dho, S. H., Kwon, H. J., Han, Y. H., Jeong, S., Kang, H. S., Lee, K. H., Rhee, S. G., and Yu, D. Y. 2003. Peroxi-redoxin II is essential for sustaining life span of erythrocytes in mice. Blood 101:5033–5038.
- Lee, C. Y., Wey, S. P., Liao, M. H., Hsu, W. L., Wu, H. Y., and Jan, T. R. 2008. A comparative study on cannabidiol-induced apoptosis in murine thymocytes and EL-4 thymoma cells. Int. Immunopharmacol. 8:732–740.
- Li, S., Ouyang, Y., Yang, G. H., and Pestka, J. J. 2000. Modulation of transcription factor AP-1 activity in murine EL-4 thymoma cells by vomitoxin (deoxynivalenol). Toxicol. Appl. Pharmacol. 163:17–25.
- Lin, J., Handschin, C., and Spiegelman, B. M. 2005. Metabolic control through the PGC-1 family of transcription co-activators. Cell Metab. 1:361–370.
- Martens, C. L., Huff, T. F., Jardieu, P., Trounstine, M. L., Coffman, R. L., Ishizaka, K., and Moore, K. W. 1985. cDNA clones encoding IgE-binding factors from a rat-mouse T-cell hybridoma. Proc. Natl. Acad. Sci. USA 82:2460–2464.
- Meiring, H. D., van Der Heeft, E., Ten Hove, G. J., and de Jong, A.P., 2002.Nanoscale LC-MS (n): Technical design and applications to peptide and protein analysis. J. Sep. Sci. 25:557–568.
- Mezghrani, A., Fassio, A., Benham, A., Simmen, T., Braakman, I., and Sitia, R. 2001. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 20:6288–6296.
- Mikami, O., Yamamoto, S., Yamanaka, N., and Nakajima, Y. 2004. Porcine hepatocyte apoptosis and reduction of albumin secretion induced by deoxynivalenol. Toxicology 204:241–249.
- Minervini, F., Fornelli, F., and Flynn, K. M. 2004. Toxicity and apoptosis induced by the mycotoxins nivalenol, deoxynivalenol, and Fumonisin B1 in a human erythroleukemia cell line. Toxicol. In Vitro 18:21–28.
- Miyamoto, S., Patel, P., and Hershey, J. W. 2005. Changes in ribosomal activity of eIF3 correlate with increased translation rates during activation of T-lymphocytes. J. Biol. Chem. 280:28251–28264.
- Moon, Y., and Pestka, J. J. 2002. Vomitoxin-induced cyclooxygenase-2 gene expression in macrophages by activation of ERK and p38 but not by JNK mitogen-activated protein kinases. Toxicol. Sci. 69:373–382.
- Naujokat, C., and Hoffman, S. 2002. Role and function of the 26 proteasome in proliferation and apoptosis. Lab. Invest. 82:965–980.
- Nishiuchi, T., Masuda, D., Nakashita, H., Ichimura, K., Shinozaki, K., Yoshida, S., Kimura, M., Yamaguchi, I., and Yamaguchi, K. 2006. Fusarium phytotoxin trichothecenes have elicitor-like activity Arbidopsis thaliana, but the activity differed among their molecular species. Mol. Plant-Microbe Interact. 19:512–520.
- Osman, A. M., van Kol, S., Peijenburg, A., Blokland, M., Pennings, J. L. A., Kleinjans, J. C. S., and van Loveren, H., 2009. Proteomic analysis of mouse thymoma EL4 cells treated with bis-(tris-n-butyltin)oxide (TBTO).J. Immunotoxicol. 6:174–183.
- Osmanagic-Myers, S. and Wiche, G., 2004. Plectin Rack 1 (receptor for activated kinase 1) scaffolding. A novel mechanism to regulate protein kinase activity. J. Biol. Chem. 279:18701–18710.
- Owen, H.R., Elser, M., Cheung, E., Gersbach, M., Kraus, W. L., and Hottiger, M. O. 2007. Mybbp1a is a novel repressor of NF-kB. J. Mol. Biol. 366:725–736.
- Pang, J. X., Ginani, N., Dongre, A. R., Hefta, S. A., and Opiteck, G. J. 2002. Biomarker discovery in urine by proteomics. J. Proteome Res. 1:161–169.
- Pestka, J. J. 2003. Deoxynivalenol-induced IgA production and IgA nephropathy-aberrant mucosal immune response with systemic repercussions. Toxicol. Lett. 140–141:287–295.
- Pestka, J. J., and Bondy, G. S. 1994. Mycotoxin-induced immunomodulation. In: Immunotoxicology and Immunopharmacology (Dean, J. H., Luster, M. I., Munson, A. E., and Kimber, I. Eds.), New York: Raven Press, pp. 163–182.
- Pestka, J. J., Zhou, H. R., Moon, Y., and Chung, Y. J. 2004. Cellular and molecular mechanisms for immune modulation by deoxynivalenol and other trichothecenes: Unraveling a paradox. Toxicol. Lett. 153:61–73.
- Pinton, P., Francesc, A., Beauchamp, E., Anne-Marie, C., Patrick, C., Francois, G., and Isabelle, P. O. 2008. Ingestion of deoxynivalenol (DON)-contaminated feed alters the pig immune response. Toxicol. Lett. 177:215–222.
- Rappsilber, J., Ryder, U., Lamond, A. I., and Mann, M., 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231–1245.
- Rocha, O., Ansari, K., and Doohan, F. M. 2005. Effects of tichothecene on eukaryotic cells: A review. Food Addit. Contam. 4:369–378.
- Shifrin, V. I., and Anderson, P. 1999. Trichothecene mycotoxins trigger a ribotoxic stress response that activates c-Jun N-terminal kinase and p38 mitogen-activated protein kinase and induces apoptosis. J. Biol. Chem. 274:13985–13992.
- Takemoto, T., Nishio, Y., Sekine, O., Ikeuchi, C., Nagai, Y., Maeno, Y., Maegawa, H., Kimura, H., and Kasiwagi, A. 2007. RBMX is a novel hepatic transcriptional regulator of SREBP-1c gene response to high-fructose diet. FEBS Lett. 581:218–222.
- Tanaka, K., Koga, Y., Taniguchi, K., and Nomoto, K. 1987. T-cell recruitment from the thymus to the spleen in tumor bearing mice: Phenotypical alteration and recruitment of thymocytes raised in a tumor bearing state. Cancer Res. 47:2136–2141.
- Tavner, F. J., Simpson, R., Tashiro, S., Favier, D., Jenkins, N. A., Gilbert, D. J., Copeland, N. G., Macmillan, J. L., Keough, R. A., Ishii, S., and Gonda, T. J. 1998. Molecular cloning reveals that the p160 Myb-binding protein is a novel, predominantly nucleolar protein which may play a role in transactivation by myb. Mol. Cell. Biol. 18:989–1002.
- Tomar, R. S., Blakley, B. R., Schiefer, H. B., and Decoteau, W. E. 1986. In vitro effects of 3-acetyl deoxynivalenol on the immune response of human peripheral blood lymphocytes. Int. J. Immunopharmac. 8:125–130.
- Tsutsumi, S., Gupta, S. K., Hogan, V., Tanaka, N., Nakamura, K.T., Nabi, I. R., and Raz, A. 2003. The enzymatic activity of phosphoglucose isomerase is not required for its cytokine function. FEBS Letts. 534:49–53.
- Tu, B. P., and Weissman, J. S. 2004. Oxidative protein folding in eukaryotes: Mechanisms and consequences. J. Cell. Biol. 164:341–346.
- Wu, Y-W., Tan, K. T., Waldmann, H., Goody, R. S., and Alexandrov, K. 2007. Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc. Natl. Acad. Sci. USA 104:12294–12299.
- Yamauchi, T., Keough, R. A., Gonda, T. J., and Ishii, S. 2008. Ribosomal stress induces processing of Mybbp1a and its translocation from the nucleolus to the nucleoplasma. Genes Cells. 13:27–39.
- Yang, G. H., Li, S., and Pestka, J. J. 2000. Down-regulation of the endoplasmic reticulum chaperone GRP78/BiP by vomitoxin (deoxynivalenol). Toxicol. Appl. Pharmacol. 162:207–217.
- Yates, J. R., Eng, J. K., McCormack, A. L., and Schieltz, D. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67:1426–1436.
- Zhou, H. R, Jia, Q., and Pestka, J. J. 2005. Ribotoxic stress response to the trichothecene deoxynivalenol in the macrophage involves the SRC family kinase HcK. Toxicol. Sci. 85:916–926.