Abstract
HLA-G is supposed to play a pivotal role in tolerance of the semi-allogeneic graft in pregnancy by inhibiting the cytotoxic functions of T and NK cells. A 14-bp insertion and/or deletion polymorphism in exon-8 has a possible role in HLA-G expression. The present study analyzed the 14-bp insertion/deletion polymorphism in normal pregnancy and recurrent miscarriage patients in order to discover a possible correlation between the 14-bp polymorphism and recurrent miscarriage (RM). In this study, genomic DNA from 200 RM patients and 200 normal fertile control individuals using the routine salting out method were isolated. Exon-8 of HLA-G gene of the two groups were amplified using polymerase chain reaction and analyzed by electrophoresis on 10% non-denaturing polyacrylamide gel electrophoresis containing ethidium bromide and visualized under ultraviolet light. HLA-G allele frequencies and genotypes in RM women and the fertile control group were compared using a Chi-square test. The results showed that there was a difference in allelic frequencies of 14-bp insertion polymorphism between fertile controls and RM patients; the frequency of +14 bp/−14 bp heterozygotes was significantly higher in RM patients as compared with fertile controls. Furthermore, the frequency of +14-bp insertion allele was significantly higher in those with RM as compared with normal fertile controls. From the findings here, it was concluded that a 14-bp insertion/deletion polymorphism in exon 8 could play a possible role in recurrent miscarriages. These results might ultimately be of significance for clinicians and those involved in understanding infertility and RM.
Introduction
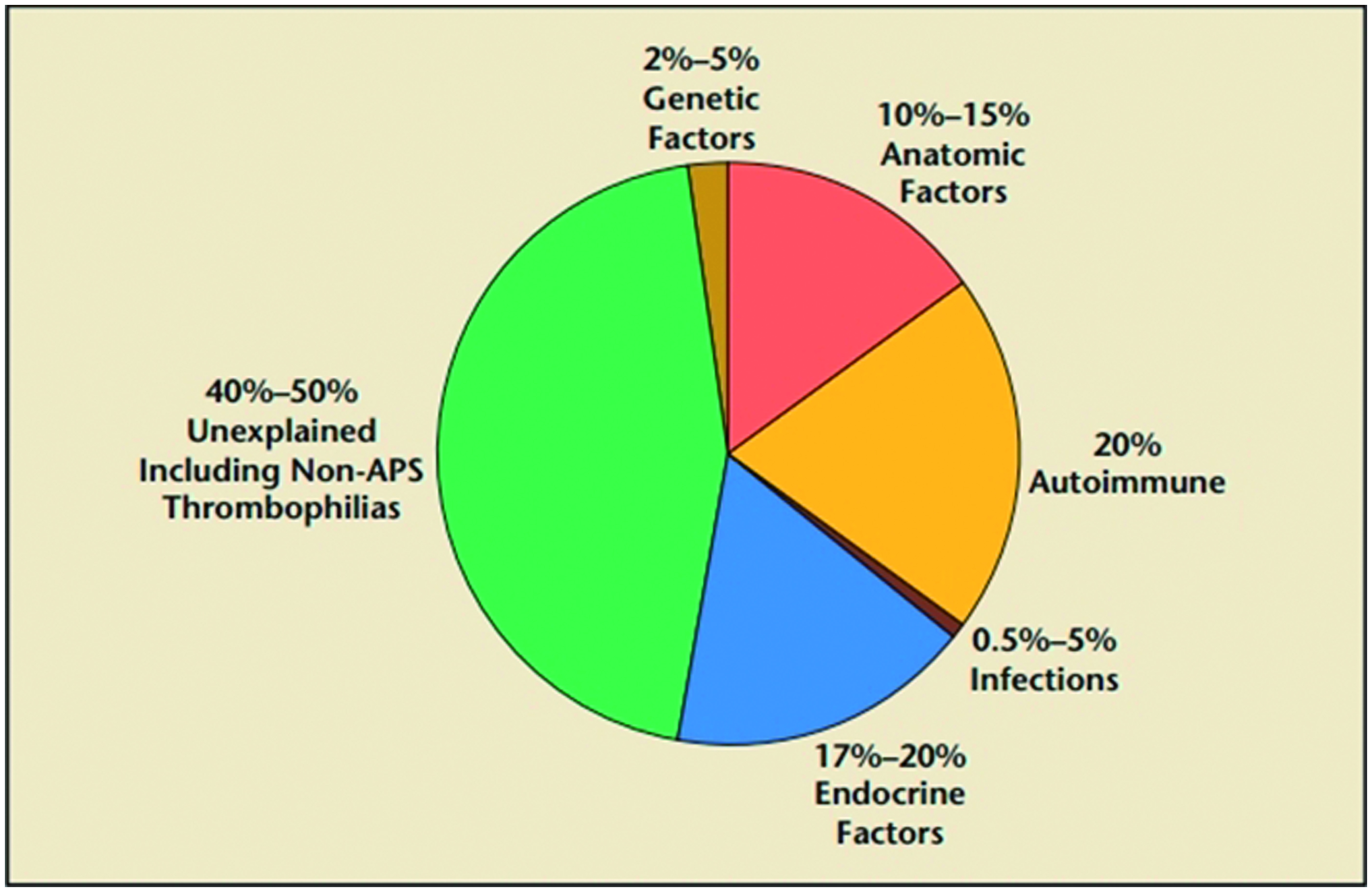
Recurrent miscarriage (RM), defined as the loss of three or more consecutive pregnancies in the first trimester of pregnancy, affects ∼1–2% of fertile women (Abdollahi et al., Citation2014; Matter & Sharif, Citation2013). At present, there are some accepted etiologies for RM (Ford & Schust, Citation2009) (), including parental chromosomal abnormalities, untreated hypo-thyroidism, uncontrolled diabetes mellitus, certain uterine anatomic abnormalities and anti-phospholipid antibody syndrome (APS), endocrine disorders, heritable/acquired thrombophilias, immunologic abnormalities, infections and environmental factors. After ruling out identifiable causes, the pathophysiology of ≈50% of RM cases remains unexplained and is, thus, termed “idiopathic” (Baek, Citation2004; Quenby et al., Citation2009; Su et al., Citation2010, Citation2011).
Figure 1. Identified and non-identified causes for RM. Information adapted from Ford and Schust (Citation2009).
Research on immunopathogenesis denotes that idiopathic RM is associated with an unbalanced materno–fetal immunological tolerance (Saini et al., Citation2011). One mechanism that circumscribes maternal immune responses at the materno–fetal interface is immunosuppression of the populations of decidual leukocytes. This leukocyte population is composed of up to 70% uterine natural killer (NK) cells and 10% T-cells. Immunosuppression of these leukocytes is believed to be mediated, in part, by the non-classical Class I human leukocyte antigen (HLA) molecule, HLA-G (Bhalla et al., Citation2006).
Genes encoding Class I HLA antigens are clustered at the telomeric end of the HLA region on chromosome 6p21. Relatively few HLA Class I genes are transcribed or translated. Expressed Class I genes are sub-divided into Class Ia, which includes HLA-A, -B and -C, and Class Ib, which includes HLA-E, -F and -G. As a non-classical major histocompatibility complex Class I molecule, HLA-G is expressed predominantly and restrictedly in extravillous trophoblast cells at the maternal–fetal interface. Alternative transcription of spliced HLA-G mRNAs encodes at least seven different HLA-G isoforms, namely the membrane-bound HLA-G1, -G2, -G3 and -G4 and the soluble HLA-G5, -G6 and -G7 proteins (Carosella et al., Citation1999; Donadi et al., Citation2011).
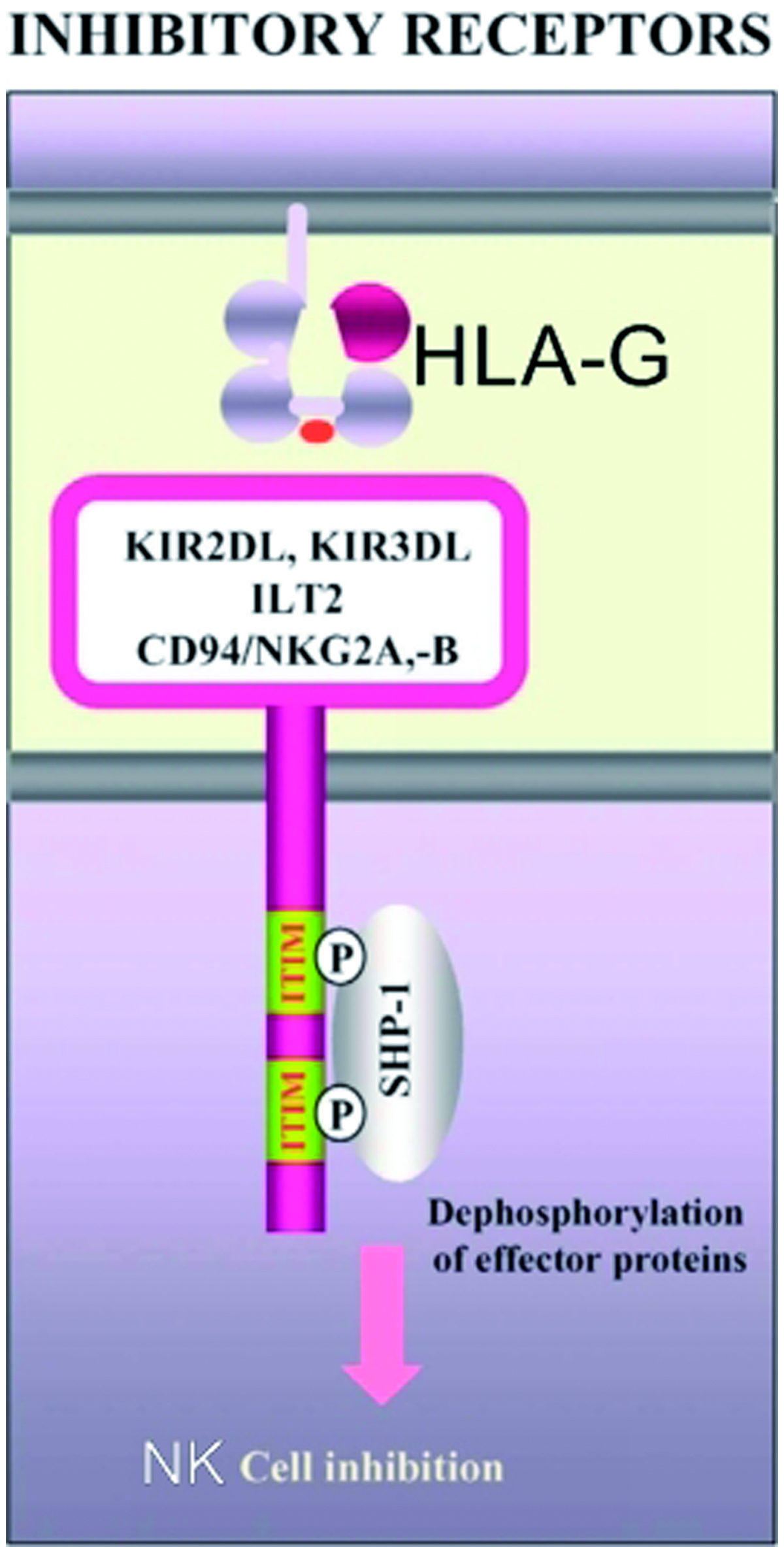
In vitro experiments showed that HLA-G may contribute to suppression of the maternal immune system; hence, a semi-allogenic fetus would be accepted during pregnancy (Tripathi et al., Citation2007). It has been shown to bind to the immunoglobulin-like transcript (ILT)-2 and killer inhibitory receptor (KIR)2DL4 on NK cells and confer protection to extravillous trophoblasts (EVT) (Bhalla et al., Citation2006) (). This points to the potential that the interaction between HLA-G and immunocompetent cells at the placental interface could be critical in determining the outcome of pregnancy. In this regard, it may be critical to look for deviations in these interactions in cases of early pregnancy disorders of unknown etiology (Emmer et al., Citation2002).
Figure 2. Inhibitory receptors associated with HLA-G. Schematic illustrates how ILT2 and KIR2DL4/HLA-G interactions can lead to inhibition of NK-cell functions.
Among its several limited polymorphisms, the 14-bp ins/del polymorphism at the 3′- untranslated region (3′UTR) of the HLA-G gene has been shown to have a consequential role in post-transcriptional regulation of HLA-G molecules (Wang et al., Citation2012). Reports have indicated that a 14-bp polymorphism was associated with HLA-G mRNA stability and isoform alternative splicing patterns (Yan et al., Citation2006); such alterations in HLA-G transcript stability influenced the levels of HLA-G expression and were thought to be associated with RM. In particular, a presence of the 14-bp insertion allele destabilizes the mRNA transcript resulting in consequently lower HLA-G expression, while the presence of the 14-bp deletion allele acts in the opposite way (Rizzo et al., Citation2012).
To provide further clarity about the potential association of these types of polymorphisms in the HLA-G gene with RM, the present study analyzed blood samples obtained from normal study subjects and patients with a history of RM. The results of these studies could then add to the knowlegde about the potential role of this specific 14-bp insertion allele in determining the outcome of a pregnancy in these and – possibly – other women outside of the particular limited group evaluated here.
Materials and methods
Study populations
In this case–control study, peripheral blood samples were obtained from women who had undergone evaluations of recurrent miscarriage (RM) at the Yazd Infertility Center over the 2013–2014 timeframe. A total of 200 patients with three or more recurrent spontaneous miscarriages (as a case group) and 200 healthy women without any history of miscarriage (as a control group) were recruited to the study; all RM and control subjects gave apprised consent for utilization of their blood. Apart from age/medical history (taken from all study subjects), the following additional data was obtained from all the RM patients: numbers of miscarriages, time of miscarriage during pregnancy and familial history of RM. Apart from their history, at the time of this study, all the women were confirmed to have normal menstrual cycles and were healthy. In addition, all of the study subjects were without anatomical, microbial, viral or genetic disease and hormone profile tests and tests for ovulation/tubal patency were normal. The Ethics Committee of Yazd Hospital as well as the University of Medical Sciences (Yazd, Iran) gave formal approval for the studies prior to the subject recruitments.
Genotyping of the 14-bp insertion/deletion polymorphisms
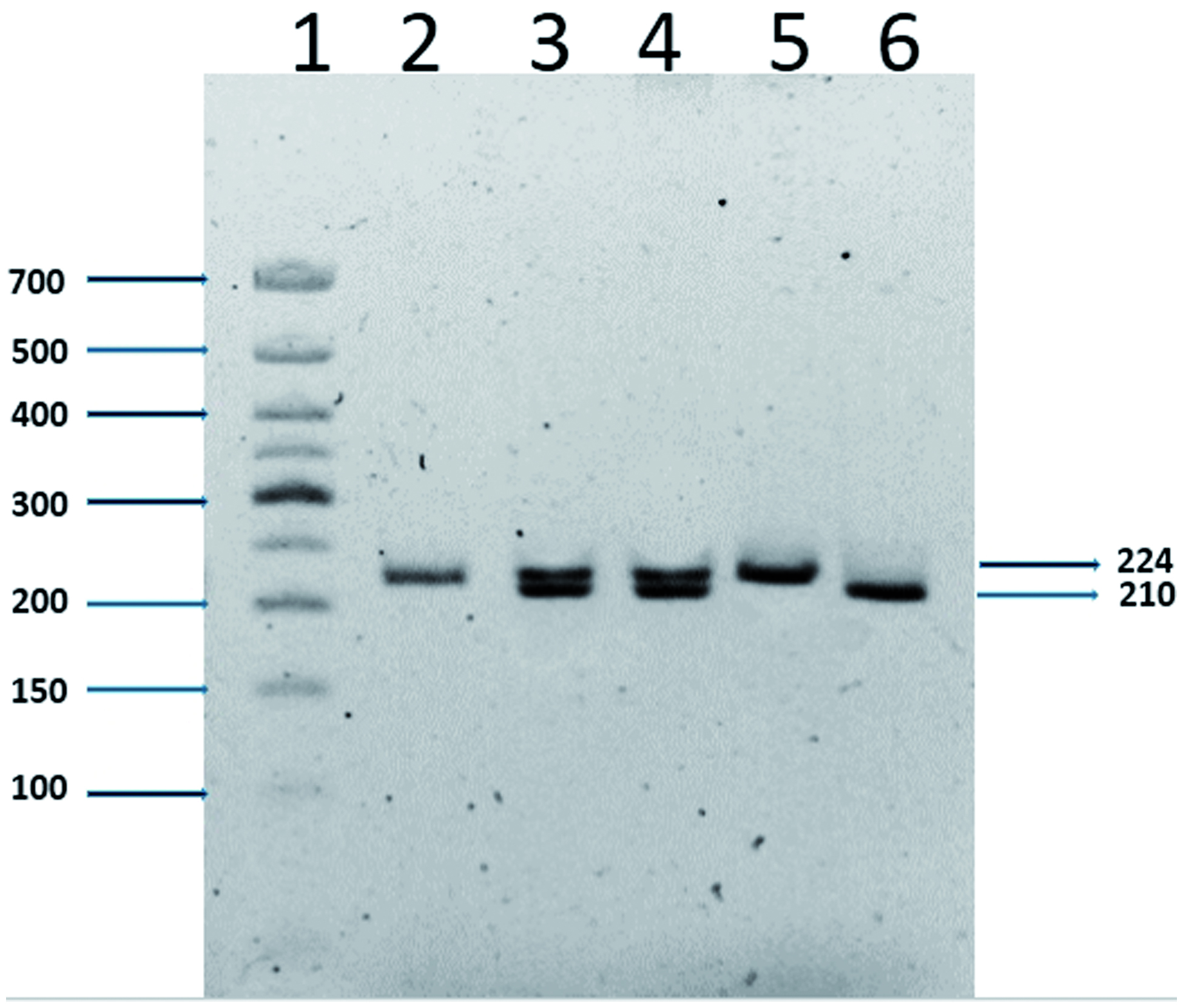
Blood samples of the controls and the patients were collected into tubes containing EDTA and molecular analyses were performed using DNA extracted from peripheral blood leukocytes after a standard salting out procedure (Nasiri et al., Citation2005). The 14-bp insertion/deletion polymorphism was genotyped using a polymerase chain reaction (PCR). Amplification was done using a forward primer 5′-GTGATGGGCTGTTTAAAGTGTCACC-3′ and a reverse primer 5′-GGAAGGAATGCAGTTCAGCATGA-3′. The cycling conditions used were: 94 °C for 5 min; 35 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 30 s and a final cycle extension at 72 °C for 5 min. All PCR products were evaluated by electrophoretic separation over 1% agarose gels. PCR products of exon 8 were analyzed via electrophoretic separation over 10% non-denaturing polyacrylamide gels containing ethidium bromide and then visualized under UV light. PCR products were of either 224 or 210 bp, respectively, depending on the insertion/deletion of the 14 bp in exon 8 ().
Figure 3. Detection of 14-bp deletion/insertion polymorphisms using electrophoresis. Lane 1: 50-bp ladder; lanes 2 and 5: homozygote for insertion; lanes 3 and 4: heterozygote; lane 6: homozygote for deletion. The figure shows a representative gel. Values to the right indicate the molecular weights of the PCR products (224 or 210 bp) depending on insertion or deletion of the 14 bp in exon 8.
Statistics
Statistical analysis was conducted using SPSS Version 19 software (SPSS, Inc., Chicago, IL). The goodness-of-fit between observed and estimated 14 bp insertion/deletion genotype frequencies was determined by a Chi-squared test. Odds ratios were calculated with a confidence interval of 95%. A difference was considered significant at a p value < 0.05.
Results
The mean age of subjects in the recurrent miscarriage study group (RM) was 35.3 years [±5.8] (range = 19–43), while the mean age in the control group was 34.9 years [±3.2] (range = 20–41) (p = 0.40). For the 19–43 year-old RM patients, there were significantly more women between 35–39 years-of-age who had experienced miscarriages (p = 0.034), so age could be an important factor in RM. Additional characteristics of the RM and control subjects are presented in .
Table 1. Clinical characteristics of patients with RM.
One purpose for collecting all of this information was to ascertain if there were possible associations of the polymorphism(s) with two important elements associated with RM, i.e. the number of miscarriages and time of miscarriage during pregnancy for the patients. Distributions of the genotype frequencies in the RM and control subjects are shown in . This study did not find a significant association between genotype and time of miscarriage during pregnancy (p = 0.866). However, as can be seen in , for the three common genotypes there were significantly more women with the heterozygous genotype who had experienced more than one (i.e. 2–5) miscarriage. Overall, the results of the current study were consistent with those reported in previous meta-analysis investigations of RM (Wang et al., Citation2013).
Figure 4. Comparison of genotype frequencies in the context of total miscarriage number. For the three common genotypes, there were significantly more women with the heterozygous genotype that experienced 2–5 miscarriages. Values shown are mean ± SD. At any of the fixed number of miscarriages, all values in the heterozygous group were significantly (p < 0.05) greater than those in either heterozygous group. For n = 200 RM subjects; actual numbers of subjects with 2, 3, 4 or 5 events were, respectively, 30, 96, 55 and 19.
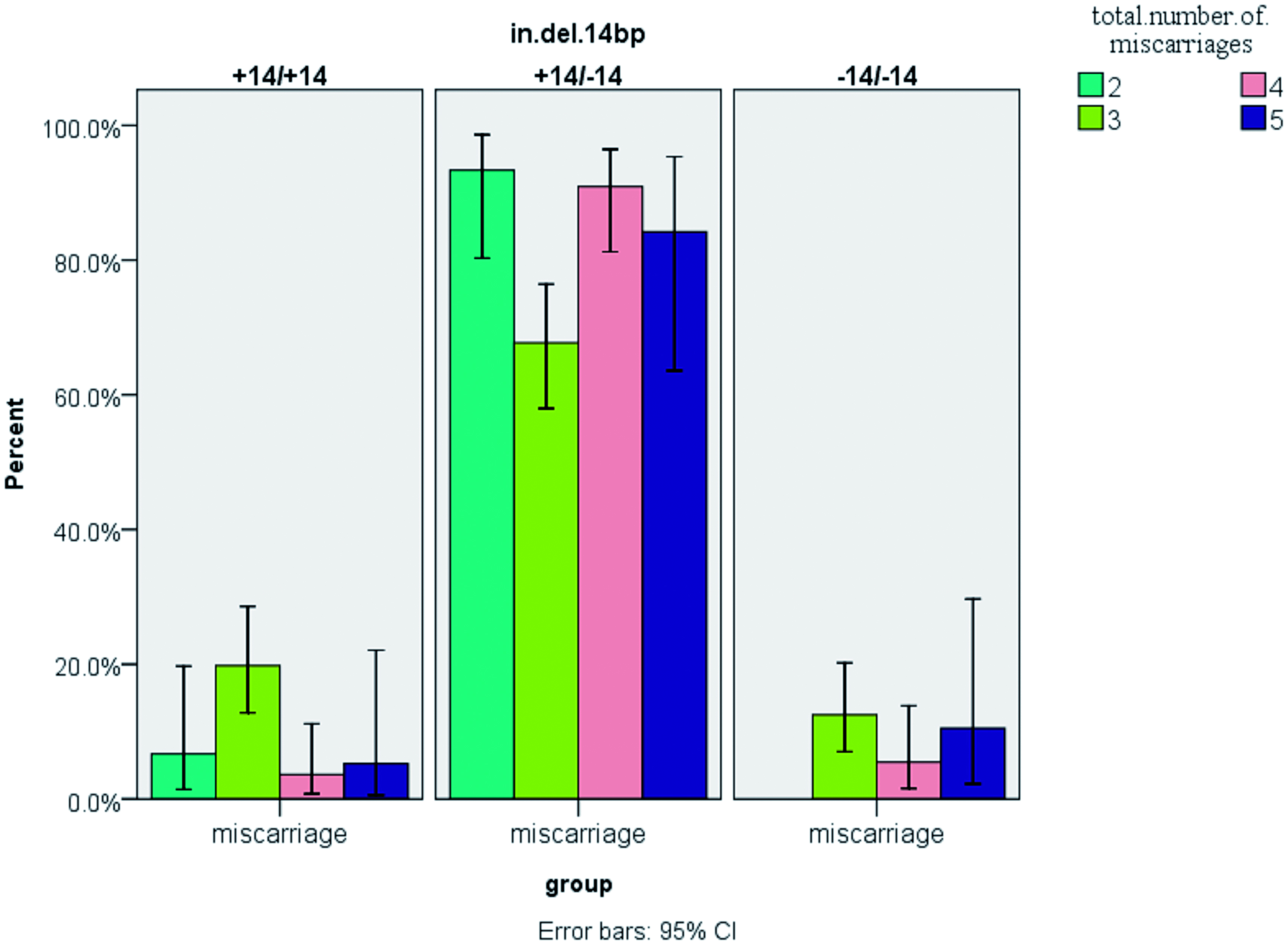
Table 2. Distribution of maternal 14-bp genotypes and alleles in control and study (RM) subjects.
The analyses also indicated the HLA-G genotype frequencies were in agreement with a Hardy-Weinberg equilibrium in this study (i.e. the frequency of +14-bp insertion and deletion alleles were, respectively, 51.75% and 48.25%, and the sum of these values = 100%). Depending on the 14-bp deletion homozygotes, 14-bp insertion homozygotes, or +14-bp /−14-bp heterozygotes of exon-8, the size of the amplified PCR products was either 224 or 210 bp or both in the case of heterozygotes. The results showed that the +14-bp /−14-bp genotype (heterozygous) frequencies were significantly different between patients with RM and fertile controls. Specifically, there were more women who were heterozygous in the RM group than among the fertile controls; however, the frequency of the +14-bp insertion allele was significantly higher in women with RM as compared to in the normal fertile controls while the genotype frequency of the −14-bp/−14-bp was significantly higher in the control women without RM (). When taken together, the data here suggested to us that there was a detectable relationship between susceptibility to RM and the presence of a +14-bp insertion allele in the HLA-G locus. Conversely, a presence of the −14-bp /−14-bp genotype appeared to be associated with some form of protective effect against recurring miscarriages.
Discussion
HLA-G has been postulated for almost two decades to be an important immunotolerizing molecule that is key in maintaining the fetal–maternal relationship (Zhu et al., Citation2010). The present study evaluated the 14-bp deletion/insertion polymorphism in the 3′ UTR of exon 8 of the HLA-G gene. This polymorphism contributes to regulation of HLA-G expression, mRNA stability and splicing patterns and, thus, may play a role in HLA-G functionality during pregnancy. The HLA-G gene encodes four membrane-bound and three soluble HLA-G isoforms as a result of alternative splicing. These proteins, also expressed on the surface of extravillous trophoblasts, are involved in inhibition of cytolytic function of uterine natural killer (NK) cells and protect fetal trophoblasts against maternal NK-cell mediated lysis. Two families of inhibitory NK receptors in humans, killer immunoglobulin-like receptors (KIR – belonging to two or three immunoglobulin [Ig] domains) and receptors known as ILT (immunoglobulin-like transcript), confer protection from NK lysis following interaction with HLA-G () (Emmer et al., Citation2002). Besides modulating the activity of NK cells, the interaction of HLA-G molecules with inhibitory receptors induces apoptosis among activated CD8+ T-cells and induces an expansion of regulatory T (Treg)-cell populations (Rizzo et al., Citation2012).
It has been reported that the 14-bp insertion allele in exon 8 of the HLA-G gene may also be associated with low levels of soluble HLA-G (sHLA-G) (Chen et al., Citation2008; Kolte et al., Citation2010). Rebmann et al. (Citation2001) reported that different variants of HLA-G were associated with variations in soluble HLA-G levels. Alleles with a 14-bp sequence deletion such as G*01041 have been designated as “high-secretor” alleles, whereas alleles G*01013 and G*0105N with the 14-bp sequence insertion were deemed “low-secretor” alleles and whose presence were associated with low levels of sHLA-G (Rebmann et al., Citation2001). Low concentrations of sHLA-G in maternal serum appeared to correlate with adverse outcomes in pregnancies; insufficient serum sHLA-G promoted attacks against trophoblasts by maternal immune cells (such as NK cells) and this could result in events common in unexplained RM, such as a loss of implantation and embryo loss (Wilczynski, Citation2006). Thus, the existence of the 14-bp insertion allele in the maternal genome may lead to subsequent pregnancy failure. The fact that the 14-bp insertion allele was previously noted to likely be a factor involved in the pathogenesis of RM (Pfeiffer et al., Citation2001) was in accordance with the results of the present study.
The results here showed there were more women that were heterozygous for the +14-bp sequence in the group with recurrent miscarriages (RM) than in fertile controls among the Iranian women studied (79.5% versus up to 57%; ). Another comparison among the three common genotypes, without considering the normal group, was carried out and the results showed that the women with the heterozygous genotype were more prone to experience a miscarriage (). The frequency of the +14-bp insertion allele showed a borderline significant increase in patients with ≥3 miscarriages while the frequency of the −14-bp /−14-bp genotype was significantly higher in the healthy fertile control women. Thus, the data suggested to us, in part, that there was a detectable relationship between susceptibility to RM and the presence of the +14-bp insertion allele in the HLA-G locus.
Only a few studies to date have reported on any potential associations of the 14-bp polymorphism with RM. A study by Tripathi et al. (Citation2004) showed that the presence of a 14-bp polymorphism itself was not a risk factor for RM, but that the number of subjects considered heterozygotes was significantly higher among those with RM. Another report by Hviid et al. (Citation2004) indicated that homozygosity for the presence of the 14-bp polymorphism was also higher among women with RM. The discrepancies between the data from the present study and that of the above studies could be partially attributable to the use of different methodologies/protocols or even the use of different ethnic populations for the analyses.
Perhaps HLA-G as an exclusive factor has a very modest effect in relation to risk for RM (Moreau et al., Citation2008). According to Yan et al. (Citation2006), a significantly different distribution of the 14-bp genotype exists among Chinese, Danish and Indian RM populations and the discrepancy may be a result of ethnic variation (see ). When compared to these other ethnic groups, several trends, however, do persist. For example, in all four groups, the incidence rate (%) of RM is seen among subjects that contained the −14 bp/+14 bp genotype. In three of the four groups, the next highest incidence was seen among those with the +14 bp/+14 bp pattern. When viewed in the context of those findings, the current data further support the hypothesis that a HLA-G polymorphism might be contributing to unexplained recurrent fetal loss in women with RM.
Table 3. Comparison of distribution of maternal 14-bp genotypes in different ethnic RM populations.
Conclusions
Based on the results here and in the context of previously reported findings in the literature, it is quite likely the 14-bp insertion allele might cause changes in the HLA-G gene that, in turn, have some significant role in determining the outcome of a pregnancy. However, since studies that have linked such polymorphisms to actual functional changes in HLA-G (such as any alterations in inhibitory activities, etc.) have not as yet been performed, any definitive linkage of a genetic change to a functional change to the ultimate induction of fetal loss must remain at this point speculative.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Acknowledgements
This work was supported by grants of Shahid Sadoughi University of Medical Sciences and National Science Foundation of Iran.
References
- Abdollahi, E., Tavasolian, F., Ghasemi, N., et al. 2014. Association between lower frequency of R381Q variant (rs11209026) in IL-23 receptor gene and increased risk of recurrent spontaneous abortion (RSA). J. Immunotoxicol. DOI: 10.3109/1547691X.2014.978056
- Baek, K. H. 2004. Aberrant gene expression associated with recurrent pregnancy loss. Mol. Human Reprod. 10:291–297
- Bhalla, A., Stone, P. R., Liddell, H. S., et al. 2006. Comparison of the expression of human leukocyte antigen (HLA)-G and HLA-E in women with normal pregnancy and those with recurrent miscarriage. Reproduction 131:583–589
- Carosella, E. D., Rouas–Freiss, N., Paul, P., and Dausset, J. 1999. HLA-G: A tolerance molecule from the major histocompatibility complex. Immunol. Today 20:60–62
- Chen, X. Y., Yan, W. H., Lin, A., et al. 2008. The 14 bp deletion polymorphisms in HLA-G gene play an important role in the expression of soluble HLA-G in plasma. Tissue Antigens 72:335–341
- Donadi, E. A., Castelli, E. C., Arnaiz-Villena, A., et al. 2011. Implications of polymorphism of HLA–G on its function, regulation, evolution and disease association. Cell. Mol. Life Sci. 68:369–395
- Emmer, P. M., Steegers, E. A., Kerstens, H. M., et al. 2002. Altered phenotype of HLA-G expressing trophoblast and decidual natural killer cells in pathological pregnancies. Human Reprod. 17:1072–1080
- Ford, H. B., and Schust, D. J. 2009. Recurrent pregnancy loss: Etiology, diagnosis, and therapy. Rev. Obstet. Gynecol. 2:76–83
- Hviid, T. V., Hylenius, S., Lindhard, A., and Christiansen, O. B. 2004. Association between human leukocyte antigen-G genotype and success of in vitro fertilization and pregnancy outcome. Tissue Antigens 64:66–69
- Kolte, A. M., Steffensen, R., Nielsen, H. S., et al. 2010. Study of structure and impact of human leukocyte antigen (HLA)-G-A, HLA-G-B, and HLA-G-DRB1 haplotypes in families with recurrent miscarriage. Human Immunol. 71:482–488
- Matter, T. F., and Sharif, F. A. 2013. HLA-G and HLA-E Gene polymorphisms in idiopathic recurrent spontaneous abortion women in Gaza Strip. Intl. J. Reprod. Contracept. Obstet. Gynecol. 2:277–283
- Moreau, P., Contu, L., Alba, F., et al. 2008. HLA-G gene polymorphism in human placentas: Possible association of G* 0106 allele with pre-eclampsia and miscarriage. Biol. Reprod. 79:459–467
- Nasiri, H., Forouzandeh, M., Rasaee, M., and Rahbarizadeh, F. 2005. Modified salting-out method: High-yield, high-quality genomic DNA extraction from whole blood using laundry detergent. J. Clin. Lab. Anal. 19:229–232
- Pfeiffer, K. A., Fimmers, R., Engels, G., et al. 2001. The HLA-G genotype is potentially associated with idiopathic recurrent spontaneous abortion. Mol. Human Reprod. 7:373–378
- Quenby, S., Nik, H., Innes, B., et al. 2009. Uterine natural killer cells and angiogenesis in recurrent reproductive failure. Human Reprod. 24:45–54
- Rebmann, V., Ven, K. V. D., Pässler, M., et al. 2001. Association Of Soluble Hla-G Plasma Levels With Hla-G Alleles. Tissue Antigens 57:15–21
- Rizzo, R., Bortolotti, D., Baricordi, O., and Fainardi, E. 2012. New insights into HLA-G and inflammatory diseases. Inflamm. Allergy Drug Targets 11:448–463
- Saini, V., Arora, S., Yadav, A., and Bhattacharjee, J. 2011. Cytokines in recurrent pregnancy loss. Clin. Chim. Acta 412:702–708
- Su, M. T., Lin, S. H., and Chen, Y. C. 2011. Genetic association studies of angiogenesis-and vasoconstriction-related genes in women with recurrent pregnancy loss: A systematic review and meta-analysis. Human Reprod. Update 17:803–812
- Su, M. T., Lin, S. H., Lee, I. W., et al. 2010. Polymorphisms of endocrine gland-derived vascular endothelial growth factor gene and its receptor genes are associated with recurrent pregnancy loss. Human Reprod. 25:2923–2930
- Tripathi, P., Abbas, A., Naik, S., and Agrawal, S. 2004. Role of 14-bp deletion in the HLA-G gene in the maintenance of pregnancy. Tissue Antigens 64:706–710
- Tripathi, P., Naik, S., and Agrawal, S. 2007. Role of HLA-G, HLA-E, and KIR2DL4 in pregnancy. Int. J. Human Genet. 7:219–233
- Wang, X., Jiang, W., and Zhang, D. 2013. Association of 14-bp insertion/deletion polymorphism of HLA-G gene with unexplained recurrent spontaneous abortion: A meta-analysis. Tissue Antigens 81:108–115
- Wang, X., Li, B., Wang, J., et al. 2012. Evidence that miR-133a causes recurrent spontaneous abortion by reducing HLA-G expression. Reprod. Biomed. Online 25:415–424
- Wilczynski, J. R. 2006. Immunological analogy between allograft rejection, recurrent abortion and pre-eclampsia: Same basic mechanism? Human Immunol. 67:492–511
- Yan, W. H., Lin, A., Chen, X., et al. 2006. Association of the maternal 14-bp insertion poly-morphism in the HLA-G gene in women with recurrent spontaneous abortions. Tissue Antigens 68:521–523
- Zhu, Y., Huo, Z., Lai, J., et al. 2010. Case-control study of a HLA-G 14-bp insertion-deletion polymorphism in women with recurrent miscarriages. Scand. J. Immunol. 71:52–54

