Abstract
Sperm nuclear proteins, the protamines (PRM) and transition nuclear proteins (TNP) play a crucial role in sperm nuclear condensation. The compact packaging of sperm DNA by protamines maintains sperm genome integrity, which is prerequisite for normal sperm function. However the effect of nucleotide variations in PRM and TNP genes on sperm DNA integrity and male fertility is not clear. This case-control study was planned to analyze PRM and TNP gene nucleotide variations and sperm DNA integrity in 100 oligozoospermic infertile men and 100 fertile controls. Protamine and TNP genes were amplified by polymerase chain reaction and sequenced. Flow cytometry-sperm chromatin structure assay (FC-SCSA) was applied to measure the DNA fragmentation index (DFI) in sperm. Semen analysis was performed as per WHO [1999] guidelines with slight modification. In total, 7 nucleotide variations including two novel changes, a non-synonymous mutation in the exon-2 of PRM2 gene (c.443C > A) and a novel insertion of T (c.396_397InsT) at the 3′ UTR region of TNP1 were detected. None of the nucleotide changes were observed with increased risk frequency in the oligozoospermic infertile men compared to the controls. Though overall DFI was significantly (p < 0.0001) higher in infertile men compared to controls (36.31 ± 7.25 vs. 26.49 ± 2.78) irrespective of nucleotide changes, no such difference was observed between 100 infertile men or pooled population of 200 with and without mutations. However it was observed that two cases with novel nucleotide changes PRM2 c.443C > A and TNP1 c.396_397InsT had higher DFI value of 34.82% and 43.85%, respectively. In conclusion, our pilot study for the first time in the Indian population revealed two rare novel mutations in sperm nuclear protein genes that are perhaps associated with higher sperm DNA fragmentation.
Introduction
Couples at higher risk for infertility are increasing in number, and majority of cases are idiopathic. Approximately half of these cases are due to male factor [Poongothai et al. Citation2009]. Men with idiopathic infertility often present with compromised semen quality. Several genetic factors such as Y chromosome genes (USP9Y, RBMY, and DAZ), androgen receptor (AR) gene, and genes regulating pituitary gonadal axis, LH and FSH are involved in the regulation of spermatogenesis, but their role in impaired semen quality is still not clearly understood [Dada et al. Citation2004; Huynh et al. Citation2002]. Spermiogenesis is a crucial event in the maturation of sperm where sperm nuclear proteins tend to play an important role in chromatin condensation. Moreover, compact packaging of sperm chromatin is essential for maintaining sperm DNA integrity, which is associated with many physiological events like sperm motility, capacitation, acrosome reaction, normal embryogenesis, and birth of healthy offspring [Castillo et al. 2010; Giwercman et al. Citation2003; Moskovtsev et al. Citation2005]. Recent studies have focused on the role of sperm nuclear proteins, the protamines (PRM) and transition nuclear proteins (TNP) on sperm function [Garcia-Peiro et al. 2011; Hammadeh et al. 2010; Hammoud et al. Citation2009; Haraguchi et al. Citation2009; Tavalaee et al. Citation2009; Tseden et al. Citation2007; Zhao et al. Citation2001]. PRM1 and PRM2 are present in equal proportions but unlike PRM1, PRM2 gene is translationally regulated in a species specific manner [Bianchi et al. 1992] and unlike TNP1, TNP2 amino acids are highly conserved among various mammals [Kremling et al. 1989]. Altered ratios of PRM1/PRM2 resulting in varied levels of chromatin condensation in sperm have also been reported as one of the important causes of male infertility [de Mateo et al. Citation2007; Mengual et al. Citation2003; Zhang et al. Citation2006]. The reason for this altered ratio may reflect the lack of post translational modification or a mutation in the PRM/TNP genes. On one hand various population studies have shown that mutations in these genes leading to infertility were not common and on the other hand, these studies have also reported novel mutations and high frequency of single nucleotide polymorphisms (SNPs) in infertile men [Adham et al. Citation2001; Carrell and Liu Citation2001; Cho et al. Citation2003; Cho et al. Citation2001; Jodar et al. 2010; Mengual, et al. Citation2003]. This pilot study is the first to report the effect of both PRM and TNP DNA sequence variations on sperm DNA integrity in the Indian population.
Results
The cDNA nucleotide positions were numbered according to their position in the genomic sequence from the start ATG codon, where A is considered as +1. In the PRM1 and PRM2 protamine genes, a total of four nucleotide changes were identified as summarized in . As summarized in , this includes one synonymous variant c.230 C > A in exon 2 of PRM1 and two variants (c.298 G > C, c.373 C > A) in intron 1 of PRM2 and one novel non-synonymus change (c. 443C > A - Gene accession ID- GU190363) in exon 2 of PRM2 as shown in . PolyPhen prediction analysis identified a novel c. 443C > A mutation as benign. An amino acid change from polar threonine to polar asparagine (p.T94N) was noted. This patient was an oligo-asthenozoospermic male with a sperm count of 17 million/ml that contained only 10% motile sperm. No difference in the frequency of PRM nucleotide changes between infertile men and controls was observed ().
Figure 1. Schematic representation of PRM1 and PRM2 gene nucleotide sequence. The blue colored regions are introns. Nucleotide changes are green and the variant is shown just above in brown with their respective position. The corresponding amino acid change is shown below the wild type in red.

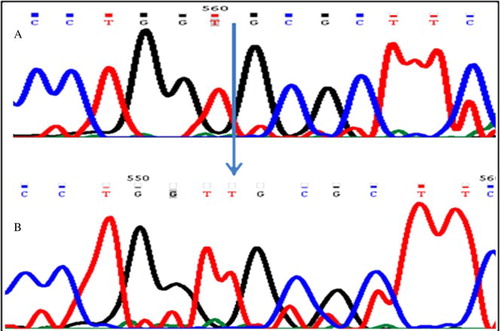
Figure 2. Sequence electrophoreogram of the SNP variant. The nucleotide variant SNP c443C > A in PRM2 gene is resolved. A) represents wild type sequence (C/C); B) represents sequence showing homozygous mutant (A/A).
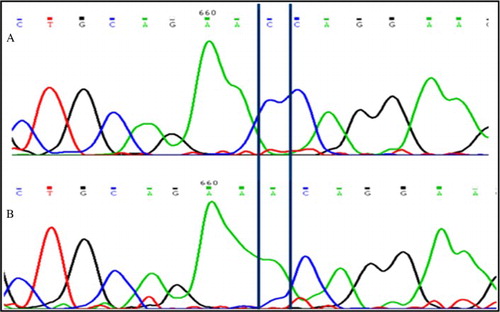
Table 1. Sperm PRM and TNP genotypic and allelic frequency in oligozoospermic infertile and control men.
In the TNP genes, a total of 3 variants and one novel insertion were identified (). A variant c.214T > C in the intron 1 of TNP1 was observed with no increase in risk frequency in the infertile men as compared to controls (). A novel insertion of nucleotide T between 396 and 397 in 3′UTR [T (c.396_397InsT) - Gene accession ID- GU190364] of TNP1 exon 1 () was identified in one patient with severe oligozoospermia (sperm count of 2.3 million/ml). Another variant c.391C > T changing amino acid polar arginine (R) to non-polar tryptophan (W) (p.R131W) in TNP2 was identified in both controls and infertile men. PolyPhen prediction analysis suggested that this change was also benign. All the nucleotide changes in the TNP gene were found with no increased risk frequency in infertile men compared to controls.
Figure 3. Schematic representations of TNP1 and TNP2 genes showing the location of the SNPs. Blue-colored regions are introns. Nucleotide changes are green and the variant is shown just above in brown with their respective position. The corresponding amino acid change is shown below the wild type in red. The non coding exon region is shown underlined.
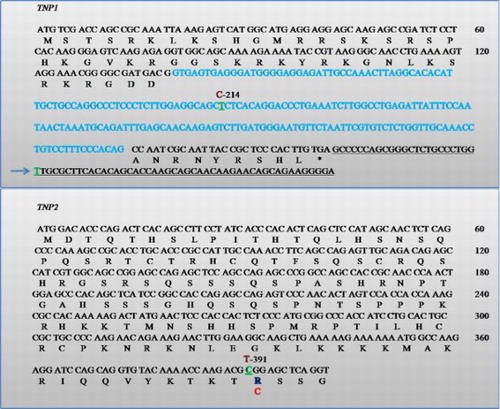
Figure 4. Sequence electrophoreogram of the SNP variant. The insertion c. Ins T 396-397 3′UTR TNP1 gene is highlighted. A) represents sequence showing homozygous wild-type (GGTG); B) represents sequence showing insertion (GGTTG).
Sperm chromatin structure assay revealed that the overall DFI in infertile men was significantly (p < 0.0001) higher as compared to controls (36.31 ± 7.25 vs. 26.49 ± 2.78). The lowest DFI in the study population was found in controls (19.82%) and the highest DFI was found in an infertile patient (43.85%). When the cases and controls were pooled together (200) and classified as with and without variations (31.27 ± 7.32 vs. 31.99 ± 5.62) no significant (p = 0.592) difference in the DFI was observed between subjects with and without variations. Similarly non significant difference (p = 0.751) in the DFI was observed between infertile men with and without nucleotide variations (36.19 ± 7.22 vs. 36.83 ± 4.57). However, a case with novel 443C > A mutation in the PRM2 gene showed a DFI - 34.82 % (), whereas the second case with novel insertion in the non-coding 3'UTR of (c.396_397InsT) TNP1 gene showed the highest DFI (43.85%) in the study ().
Discussion
Sperm has the unique function of transmitting the paternal genome to the egg. The integrity of sperm genome is a prerequisite for normal embryogenesis and fetal well being. Gradual replacement of histones by the transition proteins followed by protamines plays a crucial role in sperm nuclear condensation and maintaining nuclear DNA integrity [Lewis et al. Citation2004; Oliva Citation2006]. However, studies have shown altered PRM1/PRM2 levels in infertile men compared to normal men [Balhorn et al. Citation1988; de Yebra et al. Citation1993; Garcia-Peiro et al. 2011; Steger et al. Citation2008]. These altered levels could be either due to pathogenic mutations in the PRM and TNP genes or aberrant post translational modification of the protamines, and/or transition protein mRNAs or their products. Mutations in the PRM and TNP genes and their effect, if any, on male infertility remains controversial. However, studies are lacking in showing the effect of such mutations on sperm DNA integrity.
This study, which was the first in an Indian population has assessed both the nucleotide changes in the PRM and TNP genes as well as the corresponding changes in the DFI. In this study, the previously reported synonymous SNP c.230C > A (rs737008) in the exon 2 of the PRM1 gene was identified but with no increase in risk frequency. Other SNPs reported by Ravel et al. [2007] such as c.102G > T and c.119G > A, Aoki et al. [2006] (c. 230C > A), Gázquez et al. [2008] (c.-190 C > A), Imken et al. [2009] (c.65G > A), and Tanaka et al. [2003] (133A > G, 160C > A, 320G > A, 321C > A, 431A > G) in the PRM1gene were not identified in this population which may be due to different geographical origin and ethnicity [Aoki et al. Citation2006; Gázquez et al. Citation2008; Imken et al. Citation2009; Ravel et al. Citation2007; Tanaka et al. Citation2003]. Among the nucleotide changes observed in the PRM2 gene, the heterozygous c. 373C > A and the homozygous c.298G > C in PRM2 gene which has been previously reported were observed but with no increase in risk frequency in the study population. Moreover none of the previously reported PRM2 SNPs (−67C > T, 398G > A) and translation termination induced 248C > T were identified in this study [Imken, et al. Citation2009; Tanaka et al. Citation2003]. But, a novel non-synonymous mutation (c.443C > A) in exon 2 of PRM2 was observed in an oligo-asthenozoospermic man with a high DFI (34.82%). This is consistent with the notion that this may reflect an amino acid change that altered PRM2 protein conformation leading to deficient protamine packaging. However, this requires functional confirmation. This case had normal seminal ROS levels (data not shown) and thus oxidative stress causing DNA damage was excluded. This may require the screening of a large number of cases with functional studies to explore pathogenicity, though computational PolyPhen analysis predicted a non-pathogenic change with no alteration in the polarity of the amino acid. Therefore the PRM gene associated SNPs identified in this study which correspond to highly conserved amino acids may not contribute to male infertility in this study population.
Since TNPs are the intermediates of sperm chromatin condensation, disruption in TNP expression and binding may also impair sperm DNA integrity. Most of the studies have screened only PRM genes for SNPs [Iguchi et al. Citation2006; Kichine et al. Citation2008; Ravel et al. Citation2007] and very few have screened the TNP genes [Adham et al. Citation2001; Miyagawa et al. Citation2005]. Studies in mice have shown a lack of TNP or null mutant for TNP genes and reduced fertility [Yu et al. Citation2000; Zhao et al. Citation2004; Zhao et al. Citation2001]. In the present study, a previously reported heterozygous SNP c.214T > C (rs62180545) in intron 1 of the TNP1 gene was observed but with no increase in risk frequency of infertile when compared to the control population. However the novel insertion of T between 396 and 397 in the 3′UTR region of the TNP1 gene in a severe oligozoospermic man (sperm count-2.3 million/ml) showed a higher DFI (44%). The change in the non-coding region (5′ and 3′ UTR) may have a strong effect on protein expression due to disruption of their regulatory mechanism [Hammoud et al. Citation2007]. This mutation in 3′UTR may alter mRNA stability, localization, and translational efficiency during expression. A previously reported homozygous c.391C > T change in exon 1 of the TNP1 gene was observed at equal frequency in both infertile men and controls which is in accordance with Aoki et al. [2006]. Therefore similar to PRM genes, TNP gene nucleotide variants also had no role in disrupting spermatogenesis in this study population. Though the effect of these SNPs on the gene expression and translation is not well known, the coordinate control of PRM1, PRM2, and TNP2 genes in the protamine cluster are essential during spermatogenesis [Wykes and Krawetz Citation2003]. Similar to most of the previous reports, there is no increase in risk frequency of sperm nuclear protein gene nucleotide changes associated with male infertility in our population [Aoki et al. Citation2006; Jodar et al. 2010]. However, the two novel mutations require further screening in a large number of cases and different populations.
Infertile men with protamine deficiency may have poor sperm DNA packaging that makes the sperm genome highly susceptible to toxic environmental stimulus such as mutagenic agents, chemical and mechanical stimuli, and more susceptible to free radical induced damage and higher DNA fragmentation [Nili et al. Citation2009; Venkatesh et al. Citation2009]. When the pooled population of 200 were grouped into men with and without nucleotide changes, our results failed to support the effect of the observed mutations on sperm DNA integrity. Similar results were achieved when infertile men were grouped into men with and without mutations. However it is important to note that two cases with novel mutations c.443C > A and 3′UTR c.396_397InsT had a higher degree of sperm DNA fragmentation of DFI 34.82% and 43.85%, respectively. The DFI of the oligoasthenozoospermic case with c.443C > A is higher than the average controls (26.49%) and the effect of amino acid change on the protein structure cannot be overruled. This case also harbored the PRM1 c.230 C > A silent mutation. In the second case that presented with a novel 3′UTR c.396_397InsT in the TNP1 gene, the sperm showed the highest DFI (43.85%). The case also harbored the previously reported PRM2 intronic c.373 C > A variant. As TNP2 proteins are necessary to initiate the protamine binding to the sperm DNA, mutation in the 3'UTR may alter the mRNA stability and subsequently result in aberrant translation. Both novel variants were not observed in the control population. It would be of interest to screen these novel variants in a large number of samples. It is well known that compact packaging of sperm chromatin is essential for maintaining the sperm DNA integrity, which is essential for normal sperm motility, capacitation, acrosome reaction, successful fertilization, placentation, and embryogenesis as sperm are not mere vectors of paternal DNA but are important determinants of the developmental competence of the embryo [Wykes and Krawetz Citation2003]. Moreover, in our study the variants observed in the intronic regions such as c.298G > C, c.373C > A of PRM2 and c.391C > T, c.396_397InsT of the TNP1 gene are in the highly conserved region of eutherian mammals. Altered nucleotide changes in conserved regions may have a significant role in impaired gene expression and translation. Though protamine deficiency may lead to aberrant DNA packaging in the sperm, polymorphisms in the sperm nuclear protein gene may not have significant effect on sperm DNA integrity. An altered epigenetic mechanism and impaired post translational modification of the PRM and TNP genes should be considered to resolve the 92% of infertile men in the current study that presented with a high DFI (>30%). It is necessary to elucidate other factors involved in the maintenance of genomic integrity.
Materials and Methods
Study population
After approval from the institute ethical committee (IEC), AIIMS, informed consent was obtained from the subjects enrolled in the study. The study included 100 oligozoospermic men with an average duration of infertility of 5.60 ± 3.77 years and 100 controls (fathered a child in last one year). After thorough examination and questionnaire evaluation, oligozoospermic men (< 20 million sperm/ml) with normal 46, XY chromosomal complement, absence of Yq microdeletion, no varicocele, and no hydrocele, were only included in the study. After 4 days of sexual abstinence, semen samples were collected in a wide mouth sterile plastic container and delivered to the laboratory immediately. Semen was allowed to liquefy at room temperature and the semen parameters were analyzed as per WHO [1999] guidelines. For morphological evaluation, a 10 µl of semen smear was prepared on a clean microscope slide and fixed with 90% ethanol and stained with Giemsa. A minimum of 100 sperms per sample were evaluated for the morphological defects.
Separation of spermatozoa and sperm DNA isolation
After semen analysis, the sample was layered in isolate sperm separation medium, (Irvine Scientific Co., Santa Ana, CA, USA) and centrifuged at 300 rpm for 10 min to separate sperm cells from other cells like leukocytes, epithelial cells, and debris. After confirming the absence of other cells by examining the smear under microscope, sperm cells were washed with sperm washing media (Irvine Scientific Co.). The washed cells were subjected to DNA isolation [Laird et al. Citation1991]. Briefly, the sperm pellet was incubated at 55°C overnight with the sperm lysis medium (60 mM DTT, 4% SDS and 350µg/ml protienase K made in lysis buffer). After complete digestion, sperm DNA was precipitated by adding an equal amount of chilled isopropanol. The DNA pellet was then washed with 70% ethanol, dried at 37°C, and dissolved in Tris-EDTA (TE) buffer.
Identification of SNPs in the PRM and TNP genes
Four primer pairs were used and a 25 µl PCR reaction mixture was prepared as described by Aoki et al. [2006] with slight modification: 94°C for 4 min followed by 35 cycles of 94°C for 30s; annealing temperature for 30s [PRM1-62.5°C, PRM2-88.5°C, TNP1-55.5°C, TNP2-59°C], extension of 72°C [PRM1-60s, PRM2-80s, TNP1-50s, TNP2-70s], and a final hold for 5 min at 72°C. The amplified PCR products were confirmed by using a 2 µl aliquot of the reaction products as resolved on a 2% agarose gel. The remaining amplified products were then purified by guanidium-HCl method using a gel/PCR DNA fragments extraction kit (Geneaid, Biotech Ltd., Sijhih City, Taiwan). The purified products were directly sequenced after the samples were dissolved in 10 µl of 50% deionized formamide and sequenced in an automated DNA analyzer (Applied Biosystems, Foster City, CA, USA). The obtained sequences were analyzed using chromas software and compared with the reference sequence to identify nucleotide changes using Genebee multiple alignment (http://www.genebee.msu.su).
Sperm chromatin structure (SCSA) assay
Preparation of samples
SCSA was performed [Evenson et al. Citation2002] with slight modification. The aliquot from each ejaculate was thawed in a water bath at 37°C for 30s and diluted to a concentration of 2 × 106 sperm/ml in TNE (Tris-50mM (pH 7.5), NaCl-140mM, EDTA-5mM) buffer to a total of 200 µl in a Falcon tube. Immediately, 0.4 ml of acid detergent solution (0.08 M HCl, 0.15 M NaCl, 0.1% v/v Triton X-100, pH 1.2) was added to the Falcon tube. After exactly 30s, 1.2 ml of Acridine Orange (AO)-staining solution (6 µg AO (chromatographically purified) (Polysciences Inc. PA, USA) per ml citrate buffer (0.037 M citric acid, 0.126 M Na2HPO4, 1.1 mM EDTA disodium, 0.15 M NaCl, pH 6.0) was added. For every six test samples, one reference sample was analyzed to ensure instrument stability.
Flow cytometric measurements
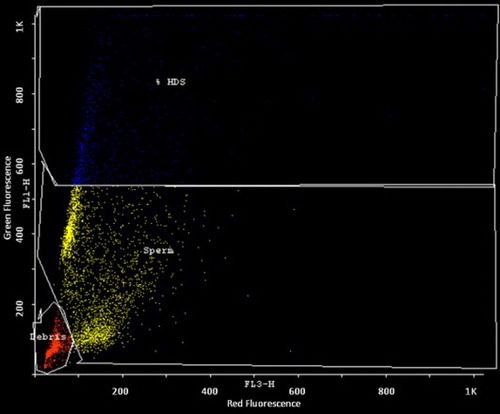
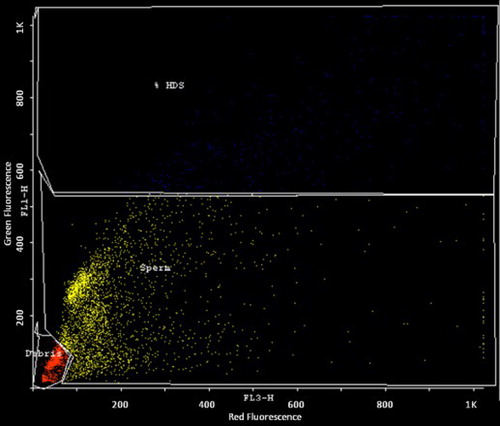
The samples were analyzed using a FAC Scan flow cytometer (BD Biosciences, CA, USA), with an air-cooled argon laser operated at 488 nm and a power of 15 mW. The green fluorescence (FL1) was collected through a 515–545 nm bandpass filter, and the red fluorescence (FL3) was collected through a 650 nm longpass filter. The sheath/sample was set on ‘low’, adjusted to a flow rate of 200 events/s when analyzing a sample containing 2 × 106 sperm/ml. Immediately after the addition of the AO staining solution, the sample was placed in the flow cytometer and run through the flow system. After complete analysis of the sample, the X-mean (red fluorescence) and Y-mean (green fluorescence) values were recorded manually after selecting gate for sperm cells using Cyflogic software version 1.2.1 (CyFlo Ltd, Finland).
DFI calculation
The sperm cells are gated after excluding debris and high DNA stainability (HDS) cells. The DNA fragmentation index (DFI) was calculated by the ratio of mean red fluorescence to the sum of mean red and mean green fluorescence. The study population including both infertile men and controls were categorized into groups with one or more mutation and without mutation. DFI was compared between these two groups to find the effect of these mutations on sperm DNA integrity.
Computational assessment of missense mutations
The homology-based program PolyPhen (polymorphism phenotyping; Division of Genetics, Department of Medicine, Brigham and Women's Hospital/Harvard Medical School, Boston, MA) was used in this study to predict the functional impact of missense mutation. PolyPhen scores of > 2.0 indicate the polymorphism is probably damaging protein function. Scores of 1.5 – 2.0 are possibly damaging, and scores of < 1.5 are likely benign.
Statistical analysis
DFI between infertile men and controls were compared by Mann-Whitney test. Genotypic and allelic frequency between infertile men and controls were analyzed by Fischer's exact test. Statistical analyses were performed using MedCalc trial version for Windows, (MedCalc Software, Mariakerke, Belgium).
Abbreviations
| PRM: | = | protamine |
| TNP: | = | transition nuclear protein |
| SCSA: | = | sperm chromatin structure assay |
| DFI: | = | DNA fragmentation index |
| UTR: | = | untranslated region |
| WHO: | = | World Health Organization. |
Acknowledgments
The authors thank all the patients and controls for their cooperation in the study.
Declaration of interest: The authors thank the Indian Council of Medical Research (ICMR) and All India Institute of Medical Sciences (AIIMS) for their partial financial support. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Adham, I.M., Nayernia, K., Burkhardt-Gottges, E., Topaloglu, O., Dixkens, C., Holstein, A.F., (2001) Teratozoospermia in mice lacking the transition protein 2 (Tnp2). Mol Hum Reprod 7:513–520.
- Aoki, V.W., Christensen, G.L., Atkins, J.F. and Carrell, D.T. (2006) Identification of novel polymorphisms in the nuclear protein genes and their relationship with human sperm protamine deficiency and severe male infertility. Fertil Steril 86:1416–1422.
- Balhorn, R., Reed, S. and Tanphaichitr, N. (1988) Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia 44:52–55.
- Bianchi, F., Rousseaux-Prevost, R., Sautiere, P. and Rousseaux, J. (1992) P2 protamines from human sperm are zinc-finger proteins with one CYS2/HIS2 motif. Biochem Biophys Res Commun 182:540–547.
- Carrell, D.T. and Liu, L. (2001) Altered protamine 2 expression is uncommon in donors of known fertility, but common among men with poor fertilizing capacity, and may reflect other abnormalities of spermiogenesis. J Androl 22:604–610.
- Castillo, J., Simon, L., de Mateo, S., Lewis, S. and Oliva, R. Protamine/DNA Ratios and DNA Damage in Native and Density Gradient Centrifugated Sperm from Infertile Patients. J Androl, Oct 21, doi:10.2164/jandrol.110.011015.
- Cho, C., Jung-Ha, H., Willis, W.D., Goulding, E.H., Stein, P., Xu, Z., (2003) Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol Reprod 69:211–217.
- Cho, C., Willis, W.D., Goulding, E.H., Jung-Ha, H., Choi, Y.C., Hecht, N.B., (2001) Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet 28:82–86.
- Dada, R., Gupta, N.P. and Kucheria, K. (2004) Yq microdeletions–azoospermia factor candidate genes and spermatogenic arrest. J Biomol Tech 15:176–183.
- de Mateo, S., Martinez-Heredia, J., Estanyol, J.M., Dominguez-Fandos, D., Vidal-Taboada, J.M., Ballesca, J.L., (2007) Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics 7:4264–4277.
- de Yebra, L., Ballesca, J.L., Vanrell, J.A., Bassas and L., Oliva, R. (1993) Complete selective absence of protamine P2 in humans. J Biol Chem 268:10553–10557.
- Evenson, D.P., Larson, K.L. and Jost, L.K. (2002) Sperm chromatin structure assay: its clinical use for detecting sperm DNA fragmentation in male infertility and comparisons with other techniques. J Androl 23:25–43.
- Garcia-Peiro, A., Martinez-Heredia, J., Oliver-Bonet, M., Abad, C., Amengual, M.J., Navarro, J., (2011) Protamine 1 to protamine 2 ratio correlates with dynamic aspects of DNA fragmentation in human sperm. Fertil Steril 95:105–109.
- Gázquez, C., Oriola, J., de Mateo, S., Vidal-Taboada, J.M., Ballesca, J.L. and Oliva, R. (2008) A common protamine 1 promoter polymorphism (-190 C->A) correlates with abnormal sperm morphology and increased protamine P1/P2 ratio in infertile patients. J Androl 29:540–548.
- Giwercman, A., Richthoff, J., Hjollund, H., Bonde, J.P., Jepson, K., Frohm, B., (2003) Correlation between sperm motility and sperm chromatin structure assay parameters. Fertil Steril 80:1404–1412.
- Hammadeh, M.E., Hamad, M.F., Montenarh, M. and Fischer-Hammadeh, C. (2010) Protamine contents and P1/P2 ratio in human spermatozoa from smokers and non-smokers. Hum Reprod 25:2708–2720.
- Hammoud, S., Emery, B.R., Aoki, V.W. and Carrell, D.T. (2007) Identification of genetic variation in the 5' and 3' non-coding regions of the protamine genes in patients with protamine deregulation. Arch Androl 53:267–274.
- Hammoud, S., Liu, L. and Carrell, D.T. (2009) Protamine ratio and the level of histone retention in sperm selected from a density gradient preparation. Andrologia 41:88–94.
- Haraguchi, T., Ishikawa, T., Yamaguchi, K. and Fujisawa, M. (2009) Cyclin and protamine as prognostic molecular marker for testicular sperm extraction in patients with azoospermia. Fertil Steril 91:1424–1426.
- Huynh, T., Mollard, R. and Trounson, A. (2002) Selected genetic factors associated with male infertility. Hum Reprod Update 8:183–198.
- Iguchi, N., Yang, S., Lamb, D.J. and Hecht, N.B. (2006) An SNP in protamine 1: a possible genetic cause of male infertility? J Med Genet 43:382–384.
- Imken, L., Rouba, H., El Houate, B., Louanjli, N., Barakat, A., Chafik, A., (2009) Mutations in the protamine locus: association with spermatogenic failure? Mol Hum Reprod 15:733–738.
- Jodar, M., Oriola, J., Mestre, G., Castillo, J., Giwercman, A., Vidal-Taboada, J.M., (2010) Polymorphisms, haplotypes and mutations in the protamine 1 and 2 genes. Int J Androl. Oct 1. doi: 10.1111/j.1365-2605.2010.01115.x.
- Kichine, E., Msaidie, S., Bokilo, A.D., Ducourneau, A., Navarro, A., Levy, N., (2008) Low-frequency protamine 1 gene transversions c.102G->T and c.-107G->C do not correlate with male infertility. J Med Genet 45:255–256.
- Kremling, H., Luerssen, H., Adham, I.M., Klemm, U., Tsaousidou, S. and Engel, W. (1989) Nucleotide sequences and expression of cDNA clones for boar and bull transition protein 1 and its evolutionary conservation in mammals. Differentiation 40:184–190.
- Laird, P.W., Zijderveld, A., Linders, K., Rudnicki, M.A., Jaenisch, R. and Berns, A. (1991) Simplified mammalian DNA isolation procedure. Nucleic Acids Res 19:4293.
- Lewis, J.D., Saperas, N., Song, Y., Zamora, M.J., Chiva, M. and Ausio, J. (2004) Histone H1 and the origin of protamines. Proc Natl Acad Sci U S A 101:4148–4152.
- Mengual, L., Ballesca, J.L., Ascaso, C. and Oliva, R. (2003) Marked differences in protamine content and P1/P2 ratios in sperm cells from percoll fractions between patients and controls. J Androl 24:438–447.
- Miyagawa, Y., Nishimura, H., Tsujimura, A., Matsuoka, Y., Matsumiya, K., Okuyama, A., (2005) Single-nucleotide polymorphisms and mutation analyses of the TNP1 and TNP2 genes of fertile and infertile human male populations. J Androl 26:779–786.
- Moskovtsev, S.I., Willis, J., Azad, A. and Mullen, J.B. (2005) Sperm DNA integrity: correlation with sperm plasma membrane integrity in semen evaluated for male infertility. Arch Androl 51:33–40.
- Nili, H.A., Mozdarani, H. and Aleyasin, A. (2009) Correlation of sperm DNA damage with protamine deficiency in Iranian subfertile men. Reprod Biomed Online 18:479–485.
- Oliva, R. (2006) Protamines and male infertility. Hum Reprod Update 12:417–435.
- Poongothai, J., Gopenath, T.S. and Manonayaki, S. (2009) Genetics of human male infertility. Singapore Med J 50:336–347.
- Ravel, C., Chantot-Bastaraud, S., El Houate, B., Berthaut, I., Verstraete, L., De Larouziere, V., (2007) Mutations in the protamine 1 gene associated with male infertility. Mol Hum Reprod 13:461–464.
- Steger, K., Wilhelm, J., Konrad, L., Stalf, T., Greb, R., Diemer, T., (2008) Both protamine-1 to protamine-2 mRNA ratio and Bcl2 mRNA content in testicular spermatids and ejaculated spermatozoa discriminate between fertile and infertile men. Hum Reprod 23:11–16.
- Tanaka, H., Miyagawa, Y., Tsujimura, A., Matsumiya, K., Okuyama, A. and Nishimune, Y. (2003) Single nucleotide polymorphisms in the protamine-1 and -2 genes of fertile and infertile human male populations. Mol Hum Reprod 9:69–73.
- Tavalaee, M., Razavi, S. and Nasr-Esfahani, M.H. (2009) Influence of sperm chromatin anomalies on assisted reproductive technology outcome. Fertil Steril 91:1119–1126.
- Tseden, K., Topaloglu, O., Meinhardt, A., Dev, A., Adham, I., Muller, C., (2007) Premature translation of transition protein 2 mRNA causes sperm abnormalities and male infertility. Mol Reprod Dev 74:273–279.
- Venkatesh, S., Deecaraman, M., Kumar, R., Shamsi, M.B. and Dada, R. (2009) Role of reactive oxygen species in the pathogenesis of mitochondrial DNA (mtDNA) mutations in male infertility. Indian J Med Res 129:127–137.
- WHO (1999) Laboratory Manual for the Examination of Human Semen and Semen-Cervical Mucus Interaction. World Health Organization. Cambridge University Press.
- Wykes, S.M. and Krawetz, S.A. (2003) Conservation of the PRM1 –> PRM2 –> TNP2 domain. DNA Seq 14:359–367.
- Yu, Y.E., Zhang, Y., Unni, E., Shirley, C.R., Deng, J.M., Russell, L.D., (2000) Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc Natl Acad Sci U S A 97:4683–4688.
- Zhang, X., San Gabriel, M. and Zini, A. (2006) Sperm nuclear histone to protamine ratio in fertile and infertile men: evidence of heterogeneous subpopulations of spermatozoa in the ejaculate. J Androl 27:414–420.
- Zhao, M., Shirley, C.R., Hayashi, S., Marcon, L., Mohapatra, B., Suganuma, R., (2004) Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis 38:200–213.
- Zhao, M., Shirley, C.R., Yu, Y.E., Mohapatra, B., Zhang, Y., Unni, E., (2001) Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol Cell Biol 21:7243–7255