Abstract
Complete deletions in the AZF (a, b, and c) sub-regions of the Y-chromosome have been shown to contribute to unexplained male infertility. However, the role of partial AZFc deletions in male infertility remains to be verified. Three types of partial AZFc deletions have been identified. They are gr/gr, b1/b3, and b2/b3 deletions. A recent meta-analysis showed that ethnic and geographical factors might contribute to the association of partial AZFc deletions with male infertility. This study analyzed the association of partial AZFc deletions in Malaysian infertile males. Fifty two oligozoospermic infertile males and 63 fertile controls were recruited to this study. Screening for partial AZFc deletions was done using the two sequence-tagged sites approach (SY1291 and SY1191) which were analyzed using both the conventional PCR gel-electrophoresis and the high resolution melt, HRM method. Gr/gr deletions were found in 11.53% of the cases and 9.52% of the controls (p = 0.725). A B2/b3 deletion was found in one of the cases (p = 0.269). No B1/b3 deletions were identified in this study. The results of HRM analysis were consistent with those obtained using the conventional PCR gel-electrophoresis method. The HRM analysis was highly repeatable (95% limit of agreement was -0.0879 to 0.0871 for SY1191 melting temperature readings). In conclusion, our study showed that partial AZFc deletions were not associated with male infertility in Malaysian subjects. HRM analysis was a reliable, repeatable, fast, cost-effective, and semi-automated method which can be used for screening of partial AZFc deletions.
Keywords:
Introduction
Unexplained male infertility can be attributed to genetic abnormalities [Dohle et al. Citation2002]. Approximatley 10-15% of cases are associated with severe male infertility [Ferlin et al. Citation2007]. Micro-deletions of the Y-chromosome azoospermia factor region (AZF) contribute significantly to the genetic causes of male infertility [O'Flynn O'Brien et al. Citation2010].
Partial deletions of the AZFc sub-regions termed g1/g1, r1/r3, and r2/r4 have been proposed as contributing to male factor infertility [Yen Citation2001] and are now abbreviated as gr/gr. Repping et al. [2003] investigated the role of the proposed partial AZFc deletions (gr/gr and b1/b3) and subsequently identified the b2/b3 deletion [Repping et al., 2004]. A recent meta-analysis [Stouffs et al. Citation2011] showed that the gr/gr deletion can increase the risk for male infertility. The increased infertility risk associated with gr/gr deletions was observed at a greater frequency in oligospermic as compared to azoopsermic European subjects. Other partial rare AZFc deletions (b2/b3 and b1/b3) are not well studied. It has been proposed that some AZFc deletions might be associated with a certain Y-chromosome lineage [Stouffs et al. Citation2011]. There has yet to be an effort to analyze the association of partial AZFc deletions in Malaysian infertile male subjects.
Conventional multiplex PCR using a panel of sequence tagged-sites (STSs) is considered as a standard screening method for AZF deletions [Simoni et al. Citation2004]. However, electrophoretic separation of the multiplex PCR products is labor-intensive only affording low resolution and sensitivity [Bor et al. Citation2003]. To overcome those issues, other technologies like real-time PCR with melt curve analysis, PCR with capillary electrophoresis and microarray technology have been used to screen for AZF deletions [Buch et al. Citation2003; Fattoruso et al. Citation2009; Zhu et al. Citation2008].
Real-time PCR-high resolution melt (HRM) analysis can be used as a rapid and inexpensive technique for various applications such as genotyping and mutation scanning [Erali et al. Citation2008; Reed et al. Citation2007] and can be adopted for multiplex-PCR [Akiyama et al. Citation2009; Erali et al. Citation2006; Seipp et al. Citation2009; Seipp et al. Citation2008]. The study described below analyzed the association of partial AZFc deletions with male infertility in Malaysian subjects and tested the possibility that these AZFc deletions might be associated with a certain Y-chromosome lineage. Furthermore, we developed and validated an HRM based method to screen for partial AZFc deletions in male infertility subjects.
Results
gr/gr partial AZFc deletions
Two cases were identified to have complete AZFc deletions and were excluded from the partial AZFc deletion analysis. shows the genotype-phenotype association of the partial AZFc deletions. Six of 52 cases were found to have the gr/gr deletions (11.53%). Six out of 63 controls were also found to have the gr/gr deletions (9.52%). Pearson's Chi Square was used to test the association of gr/gr deletions and male infertility. The Pearson's Chi-Square analysis revealed χ2 (1, N = 115) = 0.124, p = 0.725. The null hypothesis was accepted indicating that there was no association between gr/gr deletions and male infertility.
Table 1. Genotype-phenotype association of partial AZF deletions.
b2/b3 and b1/b3 partial AZFc deletions
In this study, one case (1.85%) of a b2/b3 deletion was identified. The b2/b3 deletion was also absent from the control group. In comparison the b1/b3 deletion was absent from both the cases and controls. The association of b2/b3 deletions and male infertility was assessed using the Pearson's Chi Square test. The Pearson's Chi-Square analysis revealed χ2 (1, N = 115) = 1.222, p = 0.269. The null hypothesis was accepted, indicating no association between b2/b3 deletions and male infertility.
HRM analysis screening for partial AZF deletions
To screen for partial AZFc deletions, two STSs were used in HRM analysis. The STSs were SY1191 and SY1291. shows the mean, standard deviation, and minimum and maximum melting temperatures that were found for the two STSs. The results obtained by the HRM analysis () were verified by a normalized graph (). The results were consistent with those obtained by the conventional duplex-PCR and electrophoretic analysis ().
Figure 1. Examples of real-time HRM analysis results. The figure shows an example of duplex-PCR for SY1191 and SY1291. The red curve represents a subject with SY1191 deletion, while the blue curve represents a subject with SY129 deletion. The black curve represents the NTC. All the green curves represent subjects without any deletions. HRM: high resolution melt; NTC: non-template control.
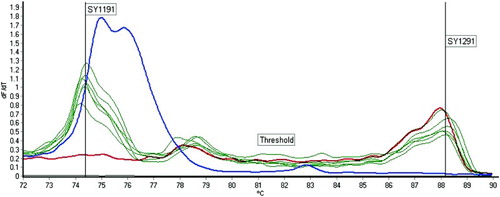
Figure 2. Normalized graph that was done to confirm the results of partial AZFc deletions. The red curve represents a sample with SY1191 deletion, the orange curve represents a sample with SY1291 deletion, the blue curves represent samples without deletions and the green curve represents the NTC. Note that the differences in the shapes of the curves confirm the deletions. NTC: non-template control.
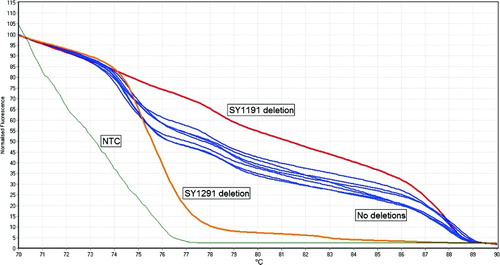
Figure 3. An example of duplex-PCR gel-electrophoresis for SY1191 and SY1291. Lane 3 had DNA of a subject with SY1191 deletion, while lane 4 had DNA of a subject with SY1291 deletion. Lanes 1 and 2 had DNA of subjects without deletions.
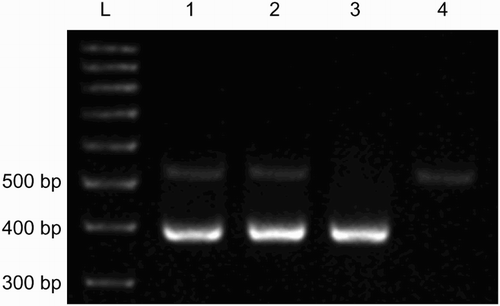
Table 2. List of the STSs that were used to screen for partial AZFc deletions with their primer concentrations, calculated melting temperatures, and calculated descriptive statistics.
Y-chromosome lineage analysis
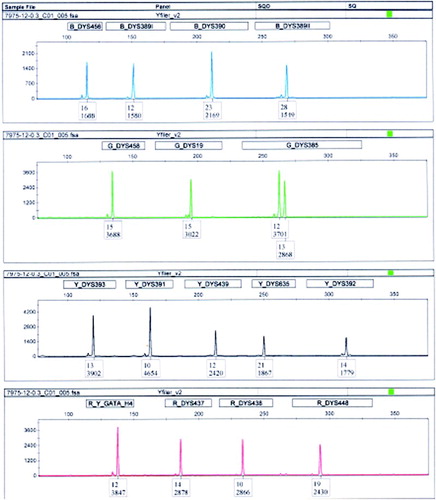
Y haplotyping was performed on seven of the cases and six of the controls that showed the gr/gr or b2/b3 deletion (). A total of 13 haplotypes was observed in the 13 samples. Statistical analysis was carried out for the haplotypes of the 12 cases of gr/gr deletions (six cases and six controls). The haplotypic richness was 5 for both the cases and the controls. The mean genetic distance between individuals was 17.29 for the cases and 18.058 for the controls. The Nei's minimum genetic distance between the cases and the controls was 0.1666. The gene diversity within each population was 0.417 for the cases and the controls. The Fst (genetic differentiation) value was 0.045 between the cases and the controls. The Y-chromosome haplogroup prediction showed that the cases were of the T (3/6), I2b1 (2/6), and L (1/6) haplogroups. In comparison the controls segregated with the following distribution into L (3/6), I2a (1/6), Q (1/6), and T (1/6) haplogroups. The single case of a b2/b3 deletion assigned to haplogroup L. The small sample size precluded the use of association statistical tests. Intra-haplogroup deletion recurrence was detected in haplogroups T, I2b1, and L.
HRM method variance
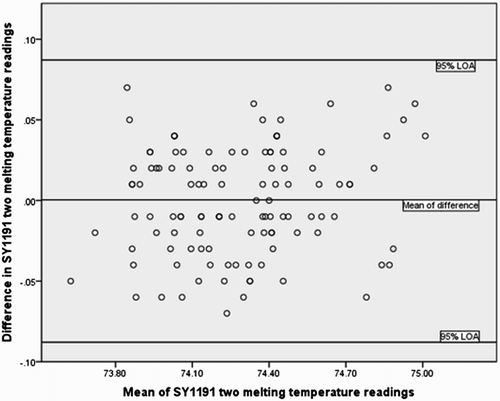
To assess reproducibility of the HRM screening method for complete and partial AZF deletions, SY254 and SY1191 melting temperatures were used respectively. Screening was repeated twice for each subject then the melting temperatures of SY254 and SY1191 were recorded. The limits of agreement (LOA) statistical method was used to test the repeatability of the HRM method [Bland and Altman Citation1986; Citation1999; Citation2007]. The standard deviation (sw) within the subjects was 0.0316. The mean of difference in the two SY254 melting temperature was 0.0004. The repeatability coefficient was 0.0875. The 95% LOA for SY1191 melting temperature readings was within a small range of -0.0879 to +0.0871. shows all study values of the difference of the mean of the two melting temperature readings bounded by 95% LOA.
Discussion
Complete AZFc deletions are well linked to male infertility. However, the significance of partial AZFc deletions (gr/gr, b2/b3, and b1/b3) in male infertility is still controversial. Ethnic and geographical factors can influence the gr/gr association with male infertility [Stouffs et al. Citation2011]. The present study was designed to determine the association of three partial AZFc deletions in Malaysian infertile male subjects.
We observed that 11.53% of cases and 9.52% of controls had gr/gr deletions but were not able to detect a significant difference in frequency of gr/gr deletions between the cases and the controls (p = 0.725). This is consistent with the data of Lu et al. [2009] that showed a frequency of 12.5% in cases and 10.2% in the controls of gr/gr deletions in Han Chinese. Lu et al. [2009] concluded that gr/gr deletions are not associated with male infertility. The frequency of the gr/gr deletions in infertile Caucasian males has been reported to be in the range of 3.2% to 5% [Giachini et al. Citation2008; Navarro-Costa et al. Citation2007; Stouffs et al. Citation2008]. The findings of this study are also in agreement with that of others [Carvalho et al. Citation2006; de Carvalho et al. Citation2006; Fernando et al. Citation2006; Hucklenbroich et al. Citation2005; Imken et al. Citation2007; Lardone et al. Citation2007; Lin et al. Citation2007; Lu et al. Citation2009; Ravel et al. Citation2009; Stouffs et al. Citation2008; Wu et al. Citation2007; Zhang et al. Citation2007] that showed no association of gr/gr deletions in infertile males. However, the results of this study differ from the findings of Repping et al. [2003] and de Llanos et al. [2005] which showed an association of gr/gr deletions in male infertility (P value of < 0.014 and 0.002, respectively). This discordance between studies may resolve as more studies confirm geographic and/or ethnic factors, e.g., perhaps gr/gr deletions are more common in Asian males so they can be found in the fertile controls as well. Hence, it is hypothesized that gr/gr deletions are a risk factor but not a cause for male infertility. Interpretation of gr/gr deletions in clinical practice must be approached with caution because the deletion can be found in fertile males as well.
This study attempted to determine whether the gr/gr deletion is inherited on a unique Y-chromosome lineage. To confirm, 13 microsatellite loci were evaluated in a proportion of gr/gr deletion–carrying cases and controls. Although microsatellite markers are not as evolutionarily stable as SNP markers, it was indicated that gr/gr deletions arose on a different Y-chromosome lineage based on the variation at these microsatellite loci. Y-haplogroup prediction showed that 50% of the cases belong to haplogroup T, while 50% of the controls belong to haplogroup L.
Some studies showed an association between b2/b3 deletions and male infertility, while other studies failed to show this association [Eloualid et al. Citation2012; Giachini et al. Citation2008; Lu et al. Citation2009; Wu et al. Citation2007; Zhang et al. Citation2007]. In the present study, only one case (1.85%) was found to present as a b2/b3 deletion. No b2/b3 deletions were observed in the controls. Together this suggests that b2/b3 deletions are not associated with male infertility (p = 0.26). On one hand, these results are in agreement with Ferlin et al. [2005], Fernando et al. [2006], and Imken et al. [2007] that failed to show an association between b2/b3 and male infertility. Similar to the present study a low frequency of b2/b3 deletions (0.29%, 1.04%, and 1.34%, respectively) in infertile males was observed. On the other hand, these results differ from some of the work of others [Eloualid et al. Citation2012; Lu et al. Citation2009; Wu et al. Citation2007] which found that b2/b3 deletions are associated with male infertility. It is difficult to reconcile these results of the b2/b3 deletion studies in Han Chinese population as on one hand they show significance [Lu et al. Citation2009; Wu et al. Citation2007] while on the other, no association [Lin et al. Citation2007] with male infertility was observed. In addition to ethnic and geographical variables, other factors like sampling methods and inclusion and exclusion criteria for the cases and controls may resolve these dicotomy. These findings suggest that, it is still too early to consider b2/b3 deletion screening in clinical practice.
Repping et al. [2003] screened 689 men and were only able to identify the b1/b3 deletion in a single Caucasian subject. The relatively low frequency has limited the observeration of b1/b3 deletions in most genetic association studies [Fernando et al. Citation2006; Giachini et al. Citation2008; Lu et al. Citation2009; Wu et al. Citation2007]. However, Shahid et al. [2011] identified 16 subjects with b1/b3 deletions in a total of 418 cases (3.82%) but did not find any b1/b3 deletions in controls in North Indian subjects. This may reflect ethnic and geographical variation. Therefore, screening for b1/b3 deletions simply remains impractical for clinical practice.
HRM analysis was used as a new screening method for both complete and partial AZF deletions. The melting temperature reading of SY1191 was used to test the repeatability of HRM screening for partial AZF deletions. The results of this study showed that all the values of the difference in the two melting temperature readings versus mean of the two melting temperature readings for SY1191 were within the 95% LOA (−0.0879 to + 0.0871). This indicates that HRM analysis for SY1191 was highly reproducible [Bland and Altman Citation1986; Citation1999; Citation2007]. The mean difference of the two melting temperature readings was 0.0004 (near zero). Small variations in the melting temperature readings were noted among samples for all STSs tested. These variations may simply refelct the differences in the DNA concentrations among the samples tested. However, these variations were very small with SD ranging from 0.2 to 0.9°C which is practically not important.
This study did not detect any differences between the results obtained by conventional multiplex PCR and those obtained with the HRM analysis method. The HRM analysis method was able to correctly identify all of the samples within the 54 cases and 63 controls. These results are consistent with the recent evaluation of HRM analysis for mutation detection by the National Genetic Reference Laboratory [White and Potts Citation2006]. This evaluation showed that Rotor-Gene 6000 (Corbett Life Sciences-QIAGEN, Hilden, Germany) had a sensitivity of 100% and a specificity ranged from 82.6 to 100% [White and Potts Citation2006].
Our study applied a new method to screen for partial AZFc deletions using real-time PCR HRM analysis. This method has a number of advantages compared with previously used technologies. HRM analysis is a rapid, probe free, simple method that can be automated [Vossen et al. Citation2009; Wittwer Citation2009]. It eliminates the labor-intensive work needed for conventional gel electrophoresis. Moreover, interpretation of the results are easier from the semi-automated genotyping ability. It is high throughput compared with previously published method of real-time PCR with melt curve analysis [Buch et al. Citation2003] and can be adapted to multiplex-PCR. In comparison to capillary electrophoresis and microarray technologies the HRM analysis is not an expensive method, as there is no post PCR separation [Vossen et al. Citation2009]. The DNA template remains intact when using the HRM method so it can be used later for sequencing or gel electrophoresis [Vossen et al. Citation2009]. AZF deletion screening was highly reproducible, sensitive, and specific. This method also offers an advantage of analysis of the cycle threshold (CT) value of the samples, which can be useful for confirmation of contamination if any peaks appear in non-template control (NTC) samples [Buch et al. Citation2003]. The findings of this study, while preliminary, suggest that HRM analysis may be further developed into a useful tool to screen for AZF deletions.
Materials and Methods
Study subjects
The ethical clearance for this study was obtained from the Ethics Committee of Faculty of Medicine and Health Sciences, Universiti Putra Malaysia (Reference number: UPM/FPSK/ PADS/T7-MJKEtikaPer/F01(JSB_MEI(09)08)). Informed consents were obtained from all the subjects included in this study. Cases subjects (infertile males) were recruited and consulted by gynecologists from the KL Fertility and Gynaecology Centre, Kuala Lumpur, Malaysia. After revision of the seminal fluid analysis results, the patients were classified into two groups. Those with total sperm count less than 20 million per mL were classified as oligozoospermic infertile males (n = 45). Those who had zero sperm count and no sperm found by testicular sperm extraction methods were classified as non-obstructive azoospermic infertile males (n = 9). Control subjects (fertile males who fathered children) were recruited from the general population (n = 63). The seminal fluid analysis was not carried out for the controls as they were selected based on proven fertility [Carvalho et al. Citation2006; de Llanos et al. Citation2005; Imken et al. Citation2007; Lardone et al. Citation2007; Lin et al. Citation2007; Navarro-Costa et al. Citation2007; Ravel et al. Citation2009; Stouffs et al. Citation2008; Wu et al. Citation2007]. Two mL of venous blood (from cubital fossa veins) was collected from the subjects enrolled in the study. DNA was extracted using a commercially available kit (Norgen Biotek Corp., Ontario, Canada). After the genomic DNA was assessed and quantified, it was stored at -20°C for further analysis.
Molecular analysis
STS analysis
Screening for AZF deletion was done using the STSs positive/negative approach. In this approach certain STSs are selected as they are uniquely present in that particular AZF sub-region. If the PCR failed to amplify certain STSs, deletion in the AZF sub-region represented by these STSs was noted. To confirm the deletion, the PCR was repeated. Screening for partial AZFc deletions (b2/b3, b1/b3, and gr/gr deletions) was done using two STSs as previously described [Giachini et al. Citation2005] (). After the revision of these STSs in the NCBI UniSTS database at http://www.ncbi.nlm.nih.gov/sites/entrez?db=unists, the primers for each of these STSs were chosen for this study.
PCR protocol
Genomic DNA was amplified by a duplex-PCR reaction. This duplex-PCR reaction was analyzed using the Rotor-Gene 6000 (Corbett Life Sciences-QIAGEN, Hilden, Germany) real-time rotary analyzer. The reaction mixture consisted of 5 µL of 2 x ImmoMix™ (Bioline Headquarters, London, UK) master mix (containing IMMOLASE™ DNA polymerase, 50 mM MgCl2, and Ultra-pure dNTPs), primers (), 1 µl (∼40 ng) of genomic DNA, 1 µL of EvaGreen™ (Biotium, Hayward, California, USA) DNA-binding fluorescent dye, and finally distilled water was added to a final volume of 25 µl reaction. Non-template control (NTC), contained ultra pure water instead of genomic DNA, was included in each PCR run as an external control to check for contamination. DNA was amplified using initial denaturation of 95°C (15 min), followed by 40 cycles of 95°C denaturation (30 s), 57.5 °C annealing (90 s), and 72°C extension (60 s), and kept at 72°C (10 min) for final extension. Amplified PCR products were analyzed using both agarose gel electrophoresis and HRM analysis.
Gel electrophoresis
Amplified duplex-PCR products were separated on a 2.5% agarose TBE gel for 50 min using GE-100 electrophoresis system (BIOER Technology Co., Hangzhou, P.R.China). After separation, agarose gel was visualized under UV light using AlphaImager™ 2200 (proteinsimple-Alpha Innotech, California, USA).
HRM analysis
HRM analysis was carried out in 0.1°C steps from 70 - 90°C. Premelt conditioning at 70°C was carried out for 90 s. The analysis was then carried in 2 s incremental steps. The HRM analysis was completed in 34 min after the PCR amplification.
To exclude the presence of contamination, the CT value of the NTC was at least 10 more than that of the subjects [Gibson et al. Citation1996]. In the melt curve analysis, a derivative plot was generated using a derivative of the fluorescence intensity versus the temperature. As the PCR amplified STSs, each sample displayed a fluorescence peak for each STS. Absence of the fluorescence peak indicates that the subject had a deletion in that STS. showed the calculated melting temperatures of all the STSs used. These melting temperatures had been verified by Singleplex-PCR reactions for those STSs. If multiple samples were analyzed together, slight variations in the melting temperatures among samples were tolerated due to variations in the template DNA concentrations. This variation can be verified by observing differences in the CT values. Automated genotyping was accoumplished using the Rotor-Gene 6000 software (Corbett Life Sciences-QIAGEN). The peaks of all samples were grouped into bins. Each bin was based on the peak values of one sample which was previously tested with conventional PCR-gel electrophoresis method and known to have an intact copy of the AZF gene. The bins were assigned to the peak values of the sample. After assigning the bins, genotypes were defined using the software based on the presence or absence of the bins. The software grouped all nearby peaks of multiple samples into bins and a peak value was then assigned to that bin. Genotyping results were displayed in the melt curve results window. The threshold was adjusted using the software to minimize the noise resulting from the non-specific fluorescence (arose from non-specific products, primer dimmers, or detector noise).
Y chromosome lineages analysis
Y-chromosome haplotype analysis was carried out to determine the Y-chromosome lineage of each gr/gr deletion. Genotyping of DNA samples for Y-chromosome haplotypes used the AmpFlSTR®Y-filer™ (Applied Biosystems, Foster City, CA, USA) at The Forensic Division, Department of Chemistry, Malaysia. PCR Amplification Kit was used for STR multiplex assay that amplifies 17 Y-STR loci (DYS19, DYS385a/b, DYS389I/II, DYS390, DYS391, DYS392, DYS393, DYS438, DYS439, DYS437, DYS448, DYS456, DYS458, DYS635 (Y GATA C4), and Y GATA H4) in a single PCR reaction. The analysis was done using male and female positive controls and reagent blank control. DNA was amplified using initial denaturation of 95°C (11 min), followed by 30 cycles of 94°C denaturation (60 s), 61°C annealing (60 s) and 72°C extension (60 s), and kept at 60°C (60 min) for final extension. Electrophoresis was done in approximately 26 min using the ABI Prism™ 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Data analysis was done using Using the GeneMapper ID v3.2 using an analytical threshold of 100RFU (). Y-chromosome haplogroups were predicted by the Y-STR values using Whit Athey's Haplogroup Predictor at http://www.hprg.com/hapest5/hapest5a/hapest5.htm?Order =num.
Statistical analysis
Data analysis employed the SPSS software (release 18.0.0, SPSS Inc., Chicago, IL, USA). Pearson's Chi-Square test was used for analysis of non-continuous variables such the presence or absence of deletions in the cases and control groups. A P value of less than 0.05 was considered to be statistically significant. To test for repeatability of the real-time PCR-HRM analysis, the LOA method was used as previously described [Bland and Altman Citation1986; Citation1999; Citation2007]. One-way analysis of variance (ANOVA) was done for the duplicate melting temperatures obtained for each subject (the readings of SY1191 were used to test for repeatability). The repeatability co-efficient was calculated as 2.77 multiplied by the with-in subject standard deviation (sw) that was obtained from the one-way ANOVA results. The 95% LOA (that tell us the limits of which 95% of the reading should lie with-in) is calculated from mean of differences (between the two readings of all the patients) - 2.77sw to mean of difference + 2.77sw [Bland and Altman Citation2007]. Statistical analysis of the haplotypes data used HAPLOTYPE SOFTWARE [Eliades and Eliades Citation2009]. This software was used to analyze the haplotypic richness, the mean genetic distance between individuals, the Nei's minimum genetic distance between the cases and the controls, the gene diversity within each population, and The Fst (genetic differentiation).
Abbreviations
| AZF: | = | azoospermia factor region of the Y-chromosome |
| STSs: | = | sequence tagged sites |
| HRM: | = | high resolution melt |
| LOA: | = | limits of agreement |
| STR: | = | short tandem repeat |
| NTC: | = | non-template control. |
Acknowledgments
The authors would like to thank all the staff of the KL Fertility and Gynaecology Centre for their assistance in recruiting the subjects for this study. The authors would also like to thank all the staff of Forensic Division of Kimia Malaysia for their help by doing the Y-STRs analysis.
Declaration of interest: The authors report no declarations of interest.
Author contributions: Designed the experiment, collected the data, analyzed the data, and drafted the manuscript: HA. Helped in data acquisition, analysis, and interpretation of results, reviewed the study for important intellectual content and edited the manuscript: RV. Interpreted the results and critically reviewed the study for important intellectual content: PI, PN, NF. All authors approved the final version of the manuscript.
References
- Akiyama, H., Nakamura, F., Yamada, C., Nakamura, K., Nakajima, O., Kawakami, H., (2009) A screening method for the detection of the 35S promoter and the nopaline synthase terminator in genetically modified organisms in a real-time multiplex polymerase chain reaction using high-resolution melting-curve analysis. Biol Pharm Bull 32:1824–1829.
- Bland, J.M. and Altman, D.G. (2007) Agreement between methods of measurement with multiple observations per individual. J Biopharm Stat 17:571–582.
- Bland, J.M. and Altman, D.G. (1999) Measuring agreement in method comparison studies. Stat Methods Med Res 8:135–160.
- Bland, J.M. and Altman, D.G. (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1:307–310.
- Bor, P., Hindkjaer, J., Kolvraa, S. and Ingerslev, H.J. (2003) A new approach for screening for Y microdeletions: capillary electrophoresis combined with fluorescent multiplex PCR. J Assist Reprod Genet 20:46–51.
- Buch, B., Galan, J.J., Lara, M., Ruiz, R., Segura, C., Real, L.M., (2003) Scanning of Y-chromosome azoospermia factors loci using real-time polymerase chain reaction and melting curve analysis. Fertil Steril 80:907–913.
- Carvalho, C.M., Zuccherato, L.W., Bastos-Rodrigues, L., Santos, F.R. and Pena, S.D. (2006) No association found between gr/gr deletions and infertility in Brazilian males. Mol Hum Reprod 12:269–273.
- de Carvalho, C.M., Zuccherato, L.W., Fujisawa, M., Shirakawa, T., Ribeiro-dos-Santos, A.K., Santos, S.E., (2006) Study of AZFc partial deletion gr/gr in fertile and infertile Japanese males. J Hum Genet 51:794–799.
- de Llanos, M., Ballesca, J.L., Gazquez, C., Margarit, E. and Oliva, R. (2005) High frequency of gr/gr chromosome Y deletions in consecutive oligospermic ICSI candidates. Hum Reprod 20:216–220.
- Dohle, G.R., Halley, D.J., Van Hemel, J.O., van den Ouwel, A.M., Pieters, M.H., Weber, R.F., (2002) Genetic risk factors in infertile men with severe oligozoospermia and azoospermia. Hum Reprod 17:13–16.
- Eliades, N.-G. and Eliades, D.G. (2009) HAPLOTYPE ANALYSIS: software for analysis of haplotypes data. Forest Genetics and Forest Tree Breeding, Georg-August University Goettingen (Germany), website: http://www.uni-goettingen.de/en/134935.html
- Eloualid, A., Rhaissi, H., Reguig, A., Bounaceur, S., El Houate, B., Abidi, O., (2012) Association of spermatogenic failure with the b2/b3 partial AZFc deletion. PLoS One 7:e34902.
- Erali, M., Pounder, J.I., Woods, G.L., Petti, C.A. and Wittwer, C.T. (2006) Multiplex single-color PCR with amplicon melting analysis for identification of Aspergillus species. Clin Chem 52:1443–1445.
- Erali, M., Voelkerding, K.V. and Wittwer, C.T. (2008) High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol 85:50–58.
- Fattoruso, O., Zarrilli, S., Coto, I., De Rosa, M., Lombardi, G. and Sacchetti, L. (2009) Prevalence of Y microdeletions in azoospermic and severe oligozoospermic men in Southern Italy: application of a rapid capillary electrophoresis method. J Endocrinol Invest 32:223–227.
- Ferlin, A., Arredi, B., Speltra, E., Cazzadore, C., Selice, R., Garolla, A., (2007) Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in Italy. J Clin Endocrinol Metab 92:762–770.
- Ferlin, A., Tessari, A., Ganz, F., Marchina, E., Barlati, S., Garolla, A., (2005) Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet 42:209–213.
- Fernando, L., Gromoll, J., Weerasooriya, T.R., Nieschlag, E. and Simoni, M. (2006) Y-chromosomal microdeletions and partial deletions of the Azoospermia Factor c (AZFc) region in normozoospermic, severe oligozoospermic and azoospermic men in Sri Lanka. Asian J Androl 8:39–44.
- Giachini, C., Guarducci, E., Longepied, G., Degl'Innocenti, S., Becherini, L., Forti, G., (2005) The gr/gr deletion(s): a new genetic test in male infertility? J Med Genet 42:497–502.
- Giachini, C., Laface, I., Guarducci, E., Balercia, G., Forti, G. and Krausz, C. (2008) Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet 124:399–410.
- Gibson, U.E., Heid, C.A. and Williams, P.M. (1996) A novel method for real time quantitative RT-PCR. Genome Res 6:995–1001.
- Hucklenbroich, K., Gromoll, J., Heinrich, M., Hohoff, C., Nieschlag, E. and Simoni, M. (2005) Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod 20:191–197.
- Imken, L., El Houate, B., Chafik, A., Nahili, H., Boulouiz, R., Abidi, O., (2007) AZF microdeletions and partial deletions of AZFc region on the Y chromosome in Moroccan men. Asian J Androl 9:674–678.
- Lardone, M.C., Parodi, D.A., Ebensperger, M., Penaloza, P., Cornejo, V., Valdevenito, R., (2007) AZFc partial deletions in Chilean men with severe spermatogenic failure. Fertil Steril 88:1318–1326.
- Lin, Y.W., Hsu, L.C., Kuo, P.L., Huang, W.J., Chiang, H.S., Yeh, S.D., (2007) Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat 28:486–494.
- Lu, C., Zhang, J., Li, Y., Xia, Y., Zhang, F., Wu, B., (2009) The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population. Hum Mol Genet 18:1122–1130.
- Navarro-Costa, P., Pereira, L., Alves, C., Gusmao, L., Proenca, C., Marques-Vidal, P., (2007) Characterizing partial AZFc deletions of the Y chromosome with amplicon-specific sequence markers. BMC Genomics 8:342.
- O'Flynn O'Brien, K.L., Varghese, A.C. and Agarwal, A. (2010) The genetic causes of male factor infertility: a review. Fertil Steril 93:1–12.
- Ravel, C., Chantot-Bastaraud, S., El Houate, B., Rouba, H., Legendre, M., Lorenco, D., (2009) Y-chromosome AZFc structural architecture and relationship to male fertility. Fertil Steril 92:1924–1933.
- Reed, G.H., Kent, J.O. and Wittwer, C.T. (2007) High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 8:597–608.
- Repping, S., Skaletsky, H., Brown, L., van Daalen, S.K., Korver, C.M., Pyntikova, T., (2003) Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet 35:247–251.
- Repping, S., van Daalen, S.K., Korver, C.M., Brown, L.G., Marszalek, J.D., Gianotten, J., (2004) A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics 83:1046–1052.
- Seipp, M.T., Durtschi, J.D., Voelkerding, K.V. and Wittwer, C.T. (2009) Multiplex amplicon genotyping by high-resolution melting. J Biomol Tech 20:160–164.
- Seipp, M.T., Pattison, D., Durtschi, J.D., Jama, M., Voelkerding, K.V. and Wittwer, C.T. (2008) Quadruplex genotyping of F5, F2, and MTHFR variants in a single closed tube by high-resolution amplicon melting. Clin Chem 54:108–115.
- Shahid, M., Dhillon, V.S., Khalil, H.S., Sexana, A. and Husain, S.A. (2011) Associations of Y-chromosome subdeletion gr/gr with the prevalence of Y-chromosome haplogroups in infertile patients. Eur J Hum Genet 19:23–29.
- Simoni, M., Bakker, E. and Krausz, C. (2004) EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletions. State of the art 2004. Int J Androl 27:240–249.
- Stouffs, K., Lissens, W., Tournaye, H. and Haentjens, P. (2011) What about gr/gr deletions and male infertility? Systematic review and meta-analysis. Hum Reprod Update 17:197–209.
- Stouffs, K., Tournaye, H., Van der Elst, J., Haentjens, P., Liebaers, I. and Lissens, W. (2008) Do we need to search for gr/gr deletions in infertile men in a clinical setting? Hum Reprod 23:1193–1199.
- Vossen, R.H., Aten, E., Roos, A. and den Dunnen, J.T. (2009) High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat 30:860–866.
- White, H. and Potts, G. (2006) Mutation scanning by high resolution melt analysis. Evaluation of RotorGene™ 6000 (Corbett Life Science), HR1™ and 384 well LightScanner™ (Idaho Technology), National Genetics Reference Laboratory (Wessex), Website: http://www.ngrl.org.uk/Wessex/ June 2006.
- Wittwer, C.T. (2009) High-resolution DNA melting analysis: advancements and limitations. Hum Mutat 30:857–859.
- Wu, B., Lu, N.X., Xia, Y.K., Gu, A.H., Lu, C.C., Wang, W., (2007) A frequent Y chromosome b2/b3 subdeletion shows strong association with male infertility in Han-Chinese population. Hum Reprod 22:1107–1113.
- Yen, P. (2001) The fragility of fertility. Nat Genet 29:243–244.
- Zhang, F., Lu, C., Li, Z., Xie, P., Xia, Y., Zhu, X., (2007) Partial deletions are associated with an increased risk of complete deletion in AZFc: a new insight into the role of partial AZFc deletions in male infertility. J Med Genet 44:437–444.
- Zhu, Y.J., Liu, S.Y., Wang, H., Wei, P. and Ding, X.P. (2008) The prevalence of azoospermia factor microdeletion on the Y chromosome of Chinese infertile men detected by multi-analyte suspension array technology. Asian J Androl 10:873–881