Abstract
Increasing evidence indicates that polymorphisms in genes relevant to spermatogenesis might modulate the efficiency of reproduction in men. Ring finger protein 8 (RNF8) and bromodomain testis-specific (BRDT) are two candidate genes associated with spermatogenesis. Here, we considered potential associations of 14 single nucleotide polymorphisms (SNPs) in RNF8 and BRDT genes in Chinese patients with non-obstructive azoospermia (NOA). We analyzed 361 men with NOA and 368 fertile controls by using Sequenom iplex technology. Our data did not reveal any variants associated with NOA susceptibility. However, we observed that rs104669 and rs195432 of RNF8 were in strong linkage disequilibrium. Haplotype analysis of the two SNPs indicated that the haplotype AC reduced the risk of NOA and the haplotype TC significantly evaluated the risk of NOA. Moreover, the RNF8 variants rs195432 (C/A p = 0.030), rs195434 (T/C p = 0.025), and rs2284922 (T/C p = 0.034) were correlated with the smaller testis volume.
Introduction
Infertility affects about 15% of couples who wish to have children. Male infertility accounts for 50% of all the infertility cases [Brugh and Lipshultz Citation2004; Hirsh Citation2003]. Non-obstructive azoospermia (NOA) is one of the infertility subtypes. During spermatogenesis, progenitor cells undergo mitotic and meiotic divisions followed by spermiogenesis yielding spermatozoa. Interruption of any of these phases can result in the failure of spermatogenesis, giving rise to NOA. Besides numerical and structural abnormalities of chromosomes [O’Flynn O’Brien et al. Citation2010], increasing evidence indicates that of spermatogenesis-related gene, polymorphisms might provide additional risk factors for the development of NOA [He et al. Citation2012; Jinam et al. Citation2013; Ruan et al. Citation2012].
The ring finger protein 8 (RNF8) gene is located on human chromosome 6p21.3. As a RING domain E3 ligase, RNF8 takes part in histone ubiquitination, which is important for a variety of biological processes [Li et al. Citation2010; Santos et al. Citation2010]. Spermatogenesis is arrested in RNF8-deficient mice [Ma et al. Citation2011] and RNF8 knockout mice fail to generate mature sperm [Santos et al. Citation2010]. Two potential mechanisms concerning the role of RNF8 in spermiogenesis were proposed. First, the histone protamine transition during spermiogenesis [Oliva Citation2006] being induced by RNF8-dependent histone ubiquitination [Lu et al. Citation2010; Ma et al. Citation2011; Rathke et al. Citation2007]. The absence of RNF8 led to incomplete chromatin restructuring resulting in male infertility [Cho et al. Citation2001; Shirley et al. Citation2004; Zhao et al. Citation2004]. Second, sex-linked genes that escaped post-meiotic silencing and activated in round spermatids and throughout spermiogenesis were RNF8-dependent [Mueller et al. Citation2008; Namekawa et al. Citation2006; Sin et al. Citation2012a,Citationb] including the Ssty gene family. Decreased expression of the Ssty gene family was correlated with a reduced number of testes sperm. This indicated that RNF8 played an essential role in activating these genes and its absence could result in infertility.
The human bromo-domain, testis specific (BRDT) gene is located on chromosome 1p22.1 and regulates a suite of previous testis-specific genes during both meiosis and post-meiotic phases of spermiogenesis [Gaucher et al. Citation2012] and is involved in chromatin remodeling through the post-meiotic phase. Adult Brdt−/− mouse testes lack post-meiotic germ cells [Dhar et al. Citation2012; Gaucher et al. Citation2012; Jacobson et al. Citation2000; Plaseski et al. Citation2012; Pivot-Pajot et al. Citation2003]. In this study, we genotyped 9 RNF8 SNPs and 5 BRDT SNPs to investigate whether genetic changes of the SNPs were risk factors for NOA.
Results and Discussion
summarizes the clinical characteristics of the 368 controls and 361 cases with NOA. The results showed that the testis volumes in NOA were significantly smaller than those in the controls (p < 0.001, both sides). There was no statistical difference of body mass index between the cases and the controls (p = 0.071).
Table 1. Clinical characteristics of the study population.
The genotype distributions of the 14 RNF8 and BRDT SNPs of interest did not deviate from the Hardy Weinberg Equilibrium (HWE) (p > 0.05) in the controls. Although we found that the T allele of rs104669 was associated with NOA (p = 0.038, odds ratio (OR) = 1.326, 95% confidence interval (CI) = 1.015–1.733, ) and the SNP rs104669 was associated with the risk of NOA in a dominant model (p = 0.045, OR = 1.376, and 95% CI = 1.007–1.880, ), both the p values were beyond statistical significance threshold after the Bonferroni correction (p > 0.05). In addition, our data revealed no significant associations of the SNPs of the BRDT gene with the risk of NOA.
Table 2. Analysis of RNF8 and BRDT polymorphisms associated with the risk of NOA.
Table 3. The genotype distributions of rs104669 in the controls and the NOA cases.
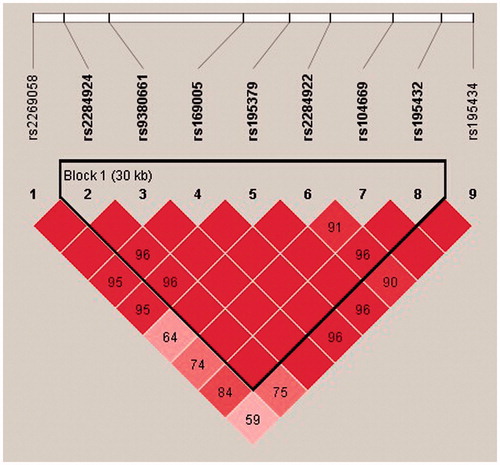
To investigate potential combinatorial effects of the RNF8 SNPs on susceptibility to NOA, we performed a linkage disequilibrium (LD) analysis. We observed that SNPs rs104669 (SNP7) and rs195432 (SNP8) were in a strong LD (). The RNF8 AC (SNP7,8) haplotype reduced the risk of NOA (p = 0.043, OR = 0.759) and the TC (SNP7,8) haplotype significantly evaluated the risk of NOA (p = 0.003, OR = 1.693). In addition, the RNF8 haplotype ATCATCTCT (SNP1-9) and the haplotype TCATCTC (SNP2-8) led to increased risk of NOA (p = 0.0086, OR = 4.052; p = 0.0085, OR = 1.630, , respectively).
Table 4. Haplotype distributions in 368 controls and 361 casesa.
We stratified the association analysis of the 14 RNF8 and BRDT SNPs as a function of testis volume and serum FSH. On one hand, RNF8 rs195432 (C > A, p = 0.03), rs195434 (T > C, p = 0.025), and rs2284922 (T > C, p = 0.034) were significantly correlated with testis volume (). On the other hand the level of serum FSH (p > 0.05, data not shown) was not correlated. We compared the relationships between the number of copies of the minor allele at the SNPs of interest and the testis volume in the NOA patients and all the samples. The result showed that there were no significant correlations between the two variables (p > 0.05, ).
Table 5. Associations of the selected polymorphisms with testis volume in the NOA patients.
Table 6. The bivariate correlation between the number of copies of the minor allele at the SNPs and testis volume in the NOA patients and all the samples.
In this study, we investigated whether RNF8 and BRDT single gene variations could be risk factors for spermatogenic failure in the Chinese population. However, we found no genetic associations between variants of RNF8 and BRDT genes and NOA. Although two independent investigations reported that the SNP rs3088232 in BRDT was associated with male infertility in the population of European descent [Aston et al. Citation2010; Plaseski et al. Citation2012], we did not recruit rs3088232 into this study because it was beyond the genetic spectrum of the Chinese population minor allele frequency (MAF = 0.022) based on the HapMap database [International HapMap Citation2005]. Small MAF would decrease statistical power for genetic analysis and likely yield a false negative.
To address the potential combinatorial effects of RNF8 SNPs on the risk of spermatogenic failure, we conducted a haplotype analysis to reveal associations that would be hidden in the evaluation of an individual SNP. We observed that RNF8 SNPs rs104669 and rs195432 were in a strong LD. NOA occurred when the Chinese individuals carried the major allele C of rs195432. The potential mechanism with regard to the effect of the RNF8 variants on spermatogenesis requires further investigation.
A significant correlation between RNF8 SNPs rs195432, rs195434, and rs2284922 with testis volume was observed (p < 0.05). However, when we compared the correlation between the number of copies of each minor allele at the SNPs (rs195432, rs195434, and rs2284922) and testis volume in the NOA patients and all the samples, we found no correlations between the two variables (p > 0.05). The reasons remained unknown. However, Rnf8−/− male mice present a reduction in testis volume [Li et al. Citation2010]. Perhaps SNPs rs195432 and rs2284922 perturb splicing, yielding other functional introns and thus alternative exons [Chamary et al. Citation2006; Chorev and Carmel Citation2012]. It was estimated that more than 50% of human genetic disorders were caused by the disruption of the normal splicing pattern [Lopez-Bigas et al. Citation2005; Wang and Cooper Citation2007]. Alternatively, the minor C allele of SNP rs2284922 is located in a conserved F-SNP (a web-based SNP analysis tool) sequence [Lee and Shatkay Citation2008]. A minor alteration of the conserved sequences could induce a phenotypic change [Venkatesh et al. Citation2011; Westerveld et al. Citation2004]. One must also consider that the variation of the SNP rs195434 in the 3′-UTR region might affect translational progress, as translational control in the 3′-untranslated region (3′-UTR) could influence protein level [de Moor et al. Citation2005]. Lastly SNP rs2284922 variation might destabilize the RNF8 mRNA [Chamary et al. Citation2006].
In conclusion, this was the first study addressing the associations of RNF8 and BRDT variants as a function of the risk of male infertility. Our data indicate that RNF8 rs104669 and rs195432 are in strong LD. RNF8 SNPs rs195432 (C/A, p = 0.03), rs195434 (T/C, p = 0.025), and rs2284922 (T/C, p = 0.034) are correlated with testis volume in the patients. Further functional analysis would be helpful to validate the biological mechanisms of RNF8 polymorphisms in the spermatogenesis failure of Chinese patients.
Materials and Methods
Ethics statement
Written informed consents for the sample collection and the subsequent research were provided by all the patients and the controls. We conducted this study according to the tenets of the Declaration of Helsinki and the ethics committee of Anhui Medical University approved this study (reference no. 2008035, 10 January 2008).
Study population
This study recruited 361 infertile men with NOA from the Reproductive Medicine Center, the First Affiliated Hospital of Anhui Medical University, China between March 2008 and July 2013. At least two semen samples from each patient were analyzed to confirm the diagnosis of azoospermia, i.e., no sperm was found after ejaculate centrifugation at 3000 × g for 10 min. All the patients’ diagnoses were based on a comprehensive andrological examination, including medical history, physical examination, semen analysis, hormone analysis, karyotype, and Y chromosome microdeletions screening. Patients with a history of orchitis, epididymitis, mono- or bilateral cryptorchidism, varicocele, hypogonadotrophic hypogonadism, obstruction/absence of vas deferens, chromosomal abnormalities, and Y microdeletions were carefully excluded from this study [Lu et al. Citation2009; Wu et al. Citation2007]. The controls consisted of 368 normozoospermia men who were in search of assisted reproduction as a result of female infertility. All the controls were drawn from the same reproductive medicine center where the patients were recruited. All patients and controls were from the Anhui province Chinese population. The age of the cases ranged from 19 to 43 and the age of the controls ranged from 20 to 53. The semen analysis for sperm concentration, motility, and morphology was performed twice following the fifth edition of the World Health Organization manuals [Cooper et al. Citation2010].
Extraction of peripheral blood DNA
Blood was collected from each subject in EDTA and stored at −80°C. Genomic DNA was extracted from peripheral blood using QIAamp DNA Blood MiDi Kit (Qiagen Inc., Hilden, Germany), according to the manufacturer’s instructions. DNA purity and concentrations were determined using a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, OH, USA) of full wave length and standardized to the concentration of 30 ng/μL.
SNP selection and genotyping
Nine RNF8 SNPs (rs104669, rs195379, rs195434, rs2284922, rs2284924, rs9380661, rs2269058, rs195432, and rs169005) and five BRDT SNPs (rs10783071, rs1156281, rs11165906, rs2183036, and rs17576636) were selected according to the HapMap database of the Chinese population [International HapMap 2005]. MAF of the SNPs were >0.05. The SNPs were genotyped using the Sequenom MassArray system at State Key Laboratory Incubation, Base of Dermatology, Ministry of National Science and Technology, Hefei, Anhui, China as described elsewhere [He et al. Citation2012]. Briefly, 15 ng of genomic DNA was used to genotype each sample. Locus-specific PCR and detection primers were designed using the MassARRAY Assay Design 3.0 software (Sequenom, San Diego, California, USA). The DNA samples were amplified by multiplex PCR, and the PCR products were then used for locus-specific single-base extension reactions. The resulting products were desalted and transferred to 384-element SpectroCHIP arrays. Allele detection was performed using MALDI-TOF MS. The mass spectrograms were analyzed using MassARRAY Typer software (Sequenom).
Data analysis
The t-test was used to compare the clinical characteristics of the controls and the cases, including the body mass index, the testicle volume on both sides, and the FSH level. The associations between the variants and the NOA risk were analyzed by calculating the OR and 95 %CI) of the MAF. HWE for each SNP was determined according to the control samples (p > 0.01). All analyses for p values were two-sided. SHEsis software (http://analysis.bio-x.cn/myAnalysis.Php) and SPSS release 17.0 statistical package were used for statistical analyses. Bonferroni correction was used to correct the p values of the allele frequency and the genotype of each SNP. For chi-square test, Fisher’s exact test was used where the number of genotype was ≤5. The haplotype analysis was performed by the haploview 4.2 software [Barrett et al. 2005]. Associations with a p value of <0.05 were considered to be of significance.
| Abbreviations | ||
| RNF8 | = | ring finger protein 8 |
| BRDT | = | bromodomain testis-specific |
| SNPs | = | single nucleotide polymorphisms |
| NOA | = | non-obstructive azoospermia |
| HWE | = | Hardy Weinberg Equilibrium |
| OR | = | odds ratio |
| LD | = | linkage disequilibrium |
| MAF | = | minor allele frequency(ies) |
Notice of Correction:
Changes have been made to Tables 2 and 4 since the original online publication date of November 6, 2014.
Declaration of interest
The authors report no conflicts of interest. This research did not receive any specific grants from any funding agencies in the public, commercial, or not-for-profit sectors. The authors alone are responsible for the content and writing of the paper.
Author contributions
Conception and design: YZ, BS, WD; Collected all samples: JR, XH; Performed the experiments: YZ, BS, XX; Acquisition of data: FZ, XZ; Analyzed the data: XZ, YZ, LY; Preparation of the manuscript: YZ, WD; Final approval of manuscript: WD, YC.
References
- Aston, K.I., Krausz. C., Laface, I., Ruiz-Castane, E., and Carrell, D.T. (2010) Evaluation of 172 candidate polymorphisms for association with oligozoospermia or azoospermia in a large cohort of men of European descent. Hum Reprod 25:1383–97
- Barrett, J.C., Fry, B., Maller, J., and Daly, M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263--5
- Brugh, V.M., 3rd and Lipshultz, L.I. (2004) Male factor infertility: evaluation and management. Med Clin North Am 88:367–85
- Chamary, J.V., Parmley, J.L., and Hurst, L.D. (2006) Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet 7:98–108
- Cho, C., Willis, W.D., Goulding, E.H., Jung-Ha, H., Choi, Y.C., Hecht, N.B., et al. (2001) Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet 28:82–6
- Chorev, M. and Carmel, L. (2012) The function of introns. Front Genet 3:1--15
- Cooper, T.G., Noonan, E., von Eckardstein, S., Auger, J., Baker, H.W., Behre, H.M., et al. (2010) World Health Organization reference values for human semen characteristics. Hum Reprod Update 16:231–45
- de Moor, C.H., Meijer, H., and Lissenden, S. (2005) Mechanisms of translational control by the 3' UTR in development and differentiation. Semin Cell Devel Biol 16:49–58
- Dhar, S., Thota, A., and Rao, M.R. (2012) Insights into role of bromodomain, testis-specific (Brdt) in acetylated histone H4-dependent chromatin remodeling in mammalian spermiogenesis. J Biol Chem 287:6387–405
- Gaucher, J., Boussouar, F., Montellier, E., Curtet, S., Buchou, T., Bertrand, S., et al. (2012) Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J 31:3809–20
- He, X.J., Ruan, J., Du, W.D., Chen, G., Zhou, Y., Xu, S., et al. (2012) PRM1 variant rs35576928 (Arg > Ser) is associated with defective spermatogenesis in the Chinese Han population. Reprod Biomed Online 25:627–34
- Hirsh, A. (2003) Male subfertility. BMJ 327:669–72
- International HapMap C (2005) A haplotype map of the human genome. Nature 437:1299–320
- Jacobson, R.H., Ladurner, A.G., King, D.S., and Tjian, R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science 288:1422–25
- Jinam, T.A., Nakaoka, H., Hosomichi, K., Mitsunaga, S., Okada, H., Tanaka, A., et al. (2013) HLA-DPB1*04:01 allele is associated with non-obstructive azoospermia in Japanese patients. Human Genet 132:1405–11
- Lee, P.H. and Shatkay, H. (2008) F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res 36:D820–4
- Li, L., Halaby, M.J., Hakem, A., Cardoso, R., El Ghamrasni, S., Harding, S., et al. (2010) Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med 207:983–97
- Lopez-Bigas, N., Audit, B., Ouzounis, C., Parra, G., and Guigo, R. (2005) Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett 579:1900–3
- Lu, C., Zhang, J., Li, Y., Xia, Y., Zhang, F., Wu, B., et al. (2009) The b2/b3 subdeletion shows higher risk of spermatogenic failure and higher frequency of complete AZFc deletion than the gr/gr subdeletion in a Chinese population. Hum Mol Genet 18:1122–30
- Lu, L.Y., Wu, J., Ye, L., Gavrilina, G.B., Saunders, T.L., and Yu, X. (2010) RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell 18:371–84
- Ma, T., Keller, J.A., and Yu, X. (2011) RNF8-dependent histone ubiquitination during DNA damage response and spermatogenesis. Acta biochimica et biophysica Sinica 43:339–45
- Mueller, J.L., Mahadevaiah, S.K., Park, P.J., Warburton, P.E., Page, D.C., and Turner, J.M. (2008) The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 40:794–9
- Namekawa, S.H., Park, P.J., Zhang, L.F., Shima, J.E., McCarrey, J.R., Griswold, M.D., et al. (2006) Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16:660–7
- O’Flynn O’Brien, K.L., Varghese, A.C., and Agarwal, A. (2010) The genetic causes of male factor infertility: a review. Fertil Steril 93:1–12
- Oliva, R. (2006) Protamines and male infertility. Hum Reprod Update 12:417–35
- Pivot-Pajot, C., Caron, C., Govin, J., Vion, A., Rousseaux, S., and Khochbin, S. (2003) Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol 23:5354–65
- Plaseski, T., Noveski, P., Popeska, Z., Efremov, G.D., and Plaseska-Karanfilska, D. (2012) Association study of single-nucleotide polymorphisms in FASLG, JMJDIA, LOC203413, TEX15, BRDT, OR2W3, INSR, and TAS2R38 genes with male infertility. J Androl 33:675–83
- Rathke, C., Baarends, W.M., Jayaramaiah-Raja, S., Bartkuhn, M., Renkawitz, R., and Renkawitz-Pohl, R. (2007) Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci 120:1689–700
- Ruan, J., He, X.J., Du, W.D., Chen, G., Zhou, Y., Xu, S., et al. (2012) Genetic variants in TEX15 gene conferred susceptibility to spermatogenic failure in the Chinese Han population. Reprod Sci 19:1190–6
- Santos, M.A., Huen, M.S., Jankovic, M., Chen, H.T., Lopez-Contreras, A.J., Klein, I.A., et al. (2010) Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J Exp Med 207:973–81
- Shirley, C.R., Hayashi, S., Mounsey, S., Yanagimachi, R., and Meistrich, M.L. (2004) Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol Reprod 71:1220–9
- Sin, H.S., Barski, A., Zhang, F., Kartashov, A.V., Nussenzweig, A., Chen, J., et al. (2012a) RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev 26:2737–48
- Sin, H.S., Ichijima, Y., Koh. E., Namiki, M., and Namekawa, S.H. (2012b) Human postmeiotic sex chromatin and its impact on sex chromosome evolution. Genome Res 22:827–36
- Venkatesh, S., Kumar, R., Deka, D., Deecaraman, M., and Dada, R. (2011) Analysis of sperm nuclear protein gene polymorphisms and DNA integrity in infertile men. Syst Biol Reprod Med 57:124–32
- Wang, G.S. and Cooper, T.A. (2007) Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8:749–61
- Westerveld, G.H., Gianotten, J., Leschot, N.J., van der Veen, F., Repping, S., and Lombardi, M.P. (2004) Heterogeneous nuclear ribonucleoprotein G-T (HNRNP G-T) mutations in men with impaired spermatogenesis. Mol Hum Reprod 10:265–9
- Wu, B., Lu, N.X., Xia, Y.K., Gu, A.H., Lu, C.C., Wang, W., et al. (2007) A frequent Y chromosome b2/b3 subdeletion shows strong association with male infertility in Han-Chinese population. Hum Reprod 22:1107–13
- Zhao, M., Shirley, C.R., Hayashi, S., Marcon, L., Mohapatra, B., Suganuma, R., et al. (2004) Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis 38:200–13