Abstract
Background and aims. Haematoloechus, digeneans parasites of amphibians, is a species-rich genus with more than 50 species around the globe. Establishing an accurate taxonomy for this group has been difficult due to high intraspecific variability. Nuclear DNA sequences have given independent information about species validity and phylogeny of the group. Materials and methods. In this paper, I test the performance of partial sequences of the mitochondrial cytochrome c oxidase subunit I (cox1) gene in the differentiation of recognized species of the genus and in the detection of potential new taxa. Samples from 13 nominal species were sequenced, plus four samples that could not be assigned to any described species based on morphology. Results. Parsimony analysis of the amplified 360 bp fragment resulted in six most parsimonious trees showing the same grouping of samples, differing in the samples' arrangement within those groups. All 13 species were recovered on the trees, and five potential new species are shown. Conclusion. Additional sampling and sequencing is necessary to support this hypothesis, but with this preliminary information the search for diagnostic characters that allow the description of the new taxa is less difficult.
Keywords::
Introduction
Species delimitation of soft-bodied animals, like helminths, can be a particularly controversial issue, especially in those cases where morphological variability can confound the taxonomy of a group (León-Règagnon et al. Citation2005), or in which morphological characters are so conservative that independent lineages cannot be differentiated (Razo-Mendivil et al. Citation2010). In these cases, DNA sequence data provide an independent source of information to differentiate diverging lineages, and to evaluate morphological characters that have been traditionally used in the taxonomy of the groups (Nadler Citation2002).
Haematoloechus, the frog lung flukes, is a species-rich genus with more than 50 described species around the globe. Morphological variability within species of this genus has caused controversy regarding species validity (Odening Citation1960; Prokopic and Krivanek Citation1974; Kennedy Citation1981). Kennedy (Citation1981), e.g. considered that only 6 out of the 15 previously known species from the USA and Canada were valid. This author concluded that morphological characters, such as the shape of the ovary and testes, arrangement of uterine loops, presence or absence of spines on body surface, sucker ratio, and egg size, exhibited intraspecific variation as the result of several factors (i.e. developmental stage, host species, crowding effect), and, therefore, they are not useful to differentiate species of Haematoloechus. Years later, using ribosomal DNA sequences (ITS2 and partial 28S), it was demonstrated that most of those morphological characters were actually useful to differentiate species, and some of them had phylogenetic value (León-Règagnon et al. Citation1999, Citation2001; León-Règagnon and Paredes-Calderón Citation2002; León-Règagnon and Brooks Citation2003). Twelve species have been reported in Mexico, six of which were named as distinct endemic species in the central region (Haematoloechus danbrooksi, Haematoloechus elongatus, Haematoloechus illimis, Haematoloechus macrorchis, Haematoloechus parcivitelarius, Haematoloechus pulcher, and Haematoloechus varioplexus), with the remaining six commonly found in southern Canada and the USA (Haematoloechus coloradensis, Haematoloechus complexus, Haematoloechus floedae, Haematoloechus longiplexus, and Haematoloechus medioplexus) (Goldberg and Bursey Citation2002; León-Règagnon Citation2003; León-Règagnon et al. Citation2005). The validity of five out of the six endemic species has been corroborated using molecular data, while morphological characters still support the validity of H. parcivitellarius (León-Règagnon Citation2003). The presence of three out of the six nearctic species in Mexico is currently recognized (H. coloradensis, H. floedae, and H. longiplexus), while records of H. medioplexus correspond to H. danbrooksi, records of Haematoloechus varioplexus are doubtful because of morphological differences observed in Mexican specimens, and records of H. complexus apparently correspond to several undescribed species according to the large genetic divergence observed in preliminary studies of Mexican populations (León-Règagnon Citation2003).
DNA barcoding (Hebert et al. Citation2003) has proven to be a useful tool to differentiate species of various taxa (e.g. Smith et al. Citation2006, Citation2008), including some groups of helminths (Ferri et al. Citation2009; Moszczynska et al. Citation2009). The aim of this study is to test the performance of partial sequences of cox1 in the discrimination of species of Haematoloechus recognized with morphology, and the detection of potential new species.
Materials and methods
Samples
For this study, we included samples of nine of the species listed above to be present in Mexico (including the doubtful H. complexus), samples of four species from other geographical regions, and four Mexican samples that could not be assigned to any described species based on morphology. Collecting localities and hosts are presented in . Worms collected from freshly killed amphibians initially were placed in saline (0.65%) for 5–10 min. For the morphological study, they were fixed by sudden immersion in hot 4% formalin and preserved in 70% ethanol. Specimens were stained with Mayer's paracarmine or Gomori's trichrome, dehydrated, cleared in methyl salicylate, and mounted in Canada balsam. Specimens were mounted permanently between cover slips and held in Cobb slides. Voucher specimen accession numbers are listed in . The following abbreviations are used: CNHE, Colección Nacional de Helmintos, Instituto de Biología, Universidad Nacional Autónoma de Mexico; USNPC, US National Parasite Collection, Beltsville, MD; and HWML, Harold W. Manter Laboratory, Lincoln, NE, USA. The following specimens were examined for comparison: H. danbrooksi León-Règagnon and Paredes-Calderón Citation2002 CNHE 4112,4151, USNPC 92220; H. floedae USNPC 30879, 84804, 091507; Haematoloechus parviplexus Irwin, 1929 USNPC 75445, 81467, HWML 20142-43, 20753, 21660, CNHE 4405; H. varioplexus Stafford, 1902 USNPC 75447, 81915, HWML 20151-20160, 38396. Specimens were assigned to a putative species in vivo using morphology and were preserved in 100% ethanol.
Table I. Hosts and collection localities of Haematoloechus species, and accession numbers of voucher specimens (in CNHE) and DNA sequences (in GenBank).
Laboratory methods
Standard phenol extraction methods were used to recover DNA from individual specimens. Laboratory protocols follow Palumbi (Citation1996) and Hillis et al. (Citation1996). PCR was used for amplifying a fragment of approximately 360 bp of the cox1 mitochondrial gene, including the 5′ end of the standard barcode (∼100 bp). Amplification and sequencing were performed using the primers JB3 5′-TTTTTTGGG CATCCTGAGGTTTAT-3′ (forward) and JB4.5 5′-TAAAGAAAGAACATAATG AAAATG-3′ (reverse) (Bowles et al. Citation1993). The amplification program consisted of 5 min at 95°C followed by 35 cycles of 30 s at 95°C, 45 s at 50°C, and 1 min 30 s at 72°C, followed by 10 min at 72°C for final elongation. PCR products were sequenced directly on an ABI Prism 310 Genetic Analyzer (Applied Biosystems, Inc., Carlsbad, California; Amersham Life Science, Inc., Buckinghamshire, England), using Thermo Sequenase radiolabeled terminator cycle sequencing kits (Amersham Life Science, Inc.). GenBank accession numbers are listed in .
Sequence analysis
Sequences were aligned manually using the computer program BioEdit (Hall Citation1999). Uncorrected distance matrices were obtained for the pairs of examined sequences and phylogenetic trees were constructed using PAUP* 4b10 (Swofford Citation2002). Unweighted parsimony analyses using branch and bound searches were performed considering character states as unordered and gaps as missing data. Nonparametric bootstrap (Felsenstein Citation1985) with 1000 pseudoreplicates was applied to evaluate the stability of nodes in the resulting topologies.
Results
A total of 37 samples from 13 recognized species were sequenced, plus four samples that could not be assigned to any described species based on morphology (). The analysis of the final alignment of 361 bp (no internal gaps) produced six most parsimonious trees (123 parsimony-informative sites; tree length = 496 steps; CI = 0.548). All of them showed the same grouping of samples with high bootstrap support. They differed in the internal arrangement of sequences within those groups, and in the position of Haematoloechus meridionalis (as sister species to H. medioplexus in four trees, ; or as sister species to H. floedae+H. illimis+Haematoloechus spp. in two trees). All 13 morphological species were differentiated on the phylogenetic trees.
Figure 1. One of six most parsimonious trees (tree length = 496; CI = 0.548) obtained from partial cox1 sequences of Haematoloechus spp. Numbers above the internode branches denote bootstrap support percentages. ★, potential new species.
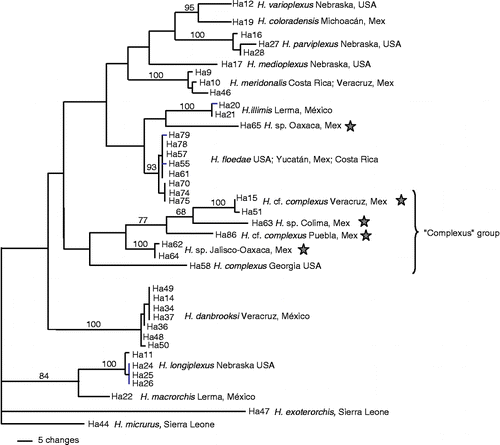
Sequence divergence did not exceed 2.2% within the same species (0.5–1.9% in H. meridionalis, 0–1.1% in H. floedae, 0–0.8% in H. danbrooksi, 0–0.5% in H. longiplexus, 0.8–2.2% in H. parviplexus, 0.5% in H. illimis), while species differed with each other by at least 6.6% (), except for H. varioplexus and H. coloradensis that differed by 2.4%. In this case, additional sequencing and reexamination of morphology are needed to clarify the specific identity of samples. The putative H. complexus was separated into three different clades (). The sample from Lithobates pipiens from Georgia, USA (collected from the type host, within the original geographical range of H. complexus [Eastern USA]) appears as an independent clade from Mexican samples, showing that H. complexus sensu stricto is not present in Mexico. Haematoloechus spp. samples Ha62, Ha63, Ha64, and Ha65 were thought to be one single species in the preliminary morphological study, but according to cox1 sequences they might belong to three different species: two of them included in the ‘complexus group’ and another that appears as sister species of H. illimis. Overall, five potentially new species of Haematoloechus were detected.
Table II. Percentage of sequence divergence among clades of Haematoloechus species recovered in this study.
Discussion
cox1 Sequences have been widely used to define species limits and phylogenetic affinities in the platyhelminthes (Bowles et al. Citation1993, Citation1995; Bowles and McManus Citation1994; Iwagami et al. Citation2000, Citation2003; Morgan et al. Citation2003; Razo-Mendivil et al. Citation2004, Citation2008); nevertheless, most of those studies used a region of the cox1 that overlaps only in a short fragment at the 5′ end of the standard barcode. The generation of primers that amplify the standard barcode region for a wide variety of platyhelminth groups has been a challenge (Moszczynska et al. Citation2009), and is still at the experimental stage. Although the cox1 fragment traditionally used in platyhelminths is shorter than the standard barcodes, it has given valuable information about the taxonomy and phylogeny of different groups, most of the time supported by other genes and morphology (Iwagami et al. Citation2000, Citation2003; Morgan et al. Citation2003; Razo-Mendivil et al. Citation2004). This is apparently also true for Haematoloechus spp., since H. complexus, which was split in several clades in our analysis, was already proposed to be a complex of ‘cryptic species’ based on ribosomal DNA sequences (León-Règagnon et al. Citation1999; León-Règagnon Citation2003; León-Règagnon and Brooks Citation2003). This split is also supported by differences in the size of the pharynx related to the size of the suckers, and the arrangement of the posterior uterine loops (personal observation). Additional sampling effort, sequencing of the complete barcode region, and combined analyses with other genes are needed to support these results. Nonetheless, even with the strongest molecular evidence for the existence of different evolutionary lineages, no formal description can be done without morphological evidence. Intraspecific morphological variability in this group renders the diagnosis of different species more difficult, but with the aid of DNA evidence we are able to identify morphological traits that can be useful to differentiate species and allow the formal description of these taxa.
Acknowledgements
The author deeply acknowledge Dan Brooks, Robert Bourgat, Valerie McKenzie, David Green, and Anindo Choudhury for donation of specimens; and Ma Antonieta Arizmendi, Rosario Mata, Elisa Cabrera, Elizabeth Martínez, Laura Paredes, Alejandro Oceguera, Rogelio Rosas, Agustín Jiménez, Ulises Razo, Gerardo Pérez, John Campbell, Eric Smith, Edmundo Pérez, and Alejandro Zaldívar for their help in field collections. Special thanks to Laura Márquez (IBUNAM) for her help in the sequencing of samples, and Luis García (CNHE), Erik Hoberg and Pat Pillitt (USNPC), and Scott Gardner (HWML) for the loan of specimens. This study was partially funded by CONACYT project 54475 to V.L.-R. and NSF grant DEB01613802 to Jonathan Campbell (University of Texas) and V.L.-R.
Declarations of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bowles J, McManus D. 1994. Genetic characterization of the Asian Taenia, a newly described Taeniid cestode of humans. Am J Trop Med Hygiene. 50:33–44.
- Bowles J, Hope M, Tiu WU, Liu X, McManus D. 1993. Nuclear and mitochondrial genetic markers highly conserved between Chinese and Philippine Schistosoma japonicum. Acta Trop. 55:217–229.
- Bowles J, Blair D, McManus D. 1995. A molecular phylogeny of the human schistosomes. Mol Phylogenet Evol. 4:103–109.
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 39:783–791.
- Ferri E, Barbuto M, Bain O, Galimberti A, Uni S, Guerrero R, Ferté H, Bandi C, Martin C, Casiraghi M. 2009. Integrated taxonomy: Traditional approach and DNA barcoding for the identification of filaroid worms and related parasites (Nematoda). Front Zool. 6:1.
- Goldberg SR, Bursey CR. 2002. Helminth parasites of seven anuran species from Northwestern Mexico. West North Am Nat. 62:160–169.
- Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc London Ser B Biol Sci. 270:313–321.
- Hillis DM, Mable BK, Moritz C. 1996. Nucleic acids IV: Sequencing and cloning. In: Hillis DM, Moritz C, Mable BK. editors. Molecular systematics. Sunderland, MA: Sinauer. p 321–383.
- Iwagami M, Ho LY, Su K, Lai PF, Fukushima M, Nakano M, Blair D, Kawashima K, Agatsuma T. 2000. Molecular phylogeographic studies on Paragonimus westermanii in Asia. J Helminthol. 74:315–322.
- Iwagami M, Monroy C, Rosas RA, Pinto MR, Guevara AR, Vieira JC, Agatsuma T. 2003. A molecular phylogeographic study based on DNA sequences from individual metacercariae of Paragonimus mexicanus from Guatemala and Ecuador. J Helminthol. 77:33–38.
- Kennedy MJ. 1981. A revision of species of the genus Haematoloechus Looss, 1899 (Trematoda: Haematoloechidae) from Canada and the United States. Canadian J Zool. 59:1836–1846.
- León-Règagnon V. 2003. Incorporating morphological and molecular data in biodiversity inventories. Parasites of leopard frogs. J Parasitol. 89:S141–S148.
- León-Règagnon V, Brooks DR. 2003. Molecular phylogeny of Haematoloechus Looss, 1899 (Digenea: Plagiorchiidae), with emphasis on north American species. J Parasitol. 89:1206–1211.
- León-Règagnon V, Paredes-Calderón EL. 2002. Haematoloechus danbrooksi n. sp. (Digenea: Plagiorchioidea), from Rana vaillanti of Los Tuxtlas, Veracruz, Mexico. J Parasitol. 88:1215–1221.
- León-Règagnon V, Brooks DR, Pérez-Ponce de León G. 1999. Differentiation of Mexican species of Haematoloechus Looss, 1899 (Digenea: Plagiorchiformes): Molecular and morphological evidence. J Parasitol. 85:935–946.
- León-Règagnon V, Brooks DR, Zelmer DA. 2001. Morphological and molecular description of Haematoloechus meridionalis n. sp. (Digenea: Plagiorchioidea: Haematoloechidae) from Rana vaillanti Brocchi of Guanacaste, Costa Rica. J Parasitol. 87:1423–1427.
- León-Règagnon V, Guillén-Hernández S, Arizmendi-Espinosa MA. 2005. Intraspecific variation of Haematoloechus floedae Harwood, 1932 (Digenea: Plagiorchiidae), from Rana spp. in North and Central America. J Parasitol. 91:915–921.
- Morgan JAT, DeJong RJ, Kazibwe F, Mkoji GM, Loker ES. 2003. A newly-identified lineage of Schistosoma. Int J Parasitol. 33:977–985.
- Moszczynska A, Locke SA, McLaughlin D, Marcogliese DJ, Crease TJ. 2009. Development of primers for the mitochondrial cytochrome c oxidase I gene in digenetic trematodes (Platyhelminthes) illustrates the challenge of barcoding parasitic helminths. Mol Ecol Res. 9:75–82.
- Nadler SA. 2002. Species delimitation and nematode biodiversity: Phylogenies rule. Nematology. 4:615–625.
- Odening K. 1960. Plagiorchiidae III. (Haematoloechinae) und Omphalometrinae. In: Mertens R, Hennig W. editors. Das Tierreich. Eine Zusammenstellung und Kennzeichnung der rezenten Tierformen. Berlin: Walter de Gruyter and Co. p 1–75.
- Palumbi SR. 1996. Nucleic acids II: The polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK. editors. Molecular systematics. Sunderland, MA: Sinauer. p 205–247.
- Prokopic J, Krivanek K. 1974. Trematodes of the genus Haematoloechus Looss, and their variability. Helminthologia. 15:779–802.
- Razo-Mendivil U, León-Règagnon V, Pérez-Ponce de León G. 2004. Description of two new species of Glypthelmins Stafford, 1905 (Digenea: Macroderoididae) in Rana spp. from Mexico, based on morphology and mtDNA and rDNA sequences. Syst Parasitol. 59:199–210.
- Razo-Mendivil U, Rosas-Valdez R, Pérez-Ponce de León G. 2008. A new cryptogonimid (Digenea) from the mayan cichlid, Cichlasoma urophthalmus (Osteichthyes: Cichlidae), in several localities of the Yucatán peninsula, Mexico. J Parasitol. 94:1371–1378.
- Razo-Mendivil U, Vázquez-Domínguez E, Rosas-Valdez R, Pérez-Ponce de León G, Nadler SA. 2010. Phylogenetic analysis of nuclear and mitochondrial DNA reveals a complex of cryptic species in Crassicutis cichlasomae (Digenea: Apocreadiidae), a parasite of Middle-American cichlids. Int J Parasitol. 40:471–486.
- Smith MA, Woodley NE, Janzen DH, Hallawachs W, Hebert PDN. 2006. DNA barcodes reveal cryptic host specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc Natl Acad Sci USA. 103:3657–3662.
- Smith MA, Poyarkov NA, Hebert PDN. 2008. CO1 DNA barcoding amphibians: Take the chance, meet the challenge. Mol Ecol Res. 8:235–246.
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4 Sunderland, MA: Sinauer.
