Abstract
We investigated the genetic distances and taxonomic status among species of Helobdella, a genus of non-blood-feeding leeches, based on mitochondrial cytochrome c oxidase subunit I sequences. Sampling included 20 specimens representing nine nominal species collected in 11 states in Mexico as well as previously published sequences of different species of Helobdella from several places. A neighbor-joining tree, as well as identification of diagnostic nucleotides, was used to suggest the presence of seven species of Helobdella in Mexico including potentially two undescribed forms.
Keywords::
Introduction
The non-blood-feeding genus Helobdella Blanchard, 1896 (Annelida: Glossiphoniidae) may be the most diverse genus of leeches including more than 50 species. Even though some species have been recorded and described in all continents (with the exception of Antarctica), South America is where the highest diversity of species is found (Ringuelet Citation1985; Sawyer Citation1986). Based on phylogenetic analyses, it has been proposed that leeches in the genus Helobdella evolved from a blood-feeding ancestor that shifted to feed on the hemolymph of mollusks and other freshwater invertebrates (Siddall and Borda Citation2003; Siddall et al. Citation2005). Sawyer (Citation1986) subdivided the genus into two main groups or series of species: a “stagnalis” series defined by the presence of a chitinous nuchal scute, including the type species for the genus Helobdella stagnalis (Linnaeus 1758); and a “triserialis” series for leeches having longitudinal stripes on the dorsal surface including Helobdella triserialis Blanchard, 1849 and related forms. Phylogenetic analyses of the group recognized the monophyly of both series (Siddall and Borda Citation2003; Siddall et al. Citation2005). However, nested within those groups were species of the genera Gloiobdella and Adaetobdella that were, subsequently, synonymized with Helobdella. In Mexico, six species of Helobdella are currently recognized (Oceguera-Figueroa and León-Règagnon Citation2005; Oceguera-Figueroa Citation2007). Two belong to the “stagnalis” series: Helobdella atli (Oceguera-Figueroa & León-Règagnon 2005) and H. stagnalis. Three species are in the “triserialis” series: Helobdella virginiae (Oceguera-Figueroa Citation2007), Helobdella conchata Caballero, 1941 and H. triserialis. Two additional species now considered junior synonyms of H. triserialis have been described for Mexico: Helobdella socimulcensis Caballero, 1932 and Helobdella moorei Caballero, 1933 (see Ringuelet Citation1981). The sixth valid species is Helobdella (Gloiobdella) elongata Caste, 1900.
Here, we reanalyze the taxonomic status of the Mexican species of Helobdella including specimens representing all of the species names recorded and described from Mexico, notwithstanding their nomenclatural validity, as well as other leech representatives from several parts of the world.
Materials and methods
Specimen collection
Twenty specimens of Helobdella were collected from 2002 to 2008, primarily from Mexico (). Specimens belonging to seven nominal species of Helobdella were collected from 11 states in Mexico, one sample was collected near Hoedspruit, South Africa and two in Washington State, USA. Specimens were collected under the scientific collecting license FAUT0056 issued to Virginia León-Règagnon. Leeches were hand-collected from submerged rocks and plants. All specimens were relaxed with the gradual addition of 70% ethanol and fixed in 96% ethanol. Voucher specimens were deposited in the Colección Nacional de Helmintos, Instituto de Biología, Universidad Nacional Autónoma de México.
Table I. Taxa, localities, and GenBank accession numbers for the COX1 sequences/catalog number (Colección Nacional de Helmintos [CNHE], UNAM, México) of leeches of Helobdella spp. used in the neighbor-joining analyses.
Sequences of the mitochondrial cytochrome c oxidase subunit I (COX1) of 20 specimens from Mexico, two from the USA, and one from South Africa were newly generated for the present study. Methods of leech DNA extraction, COX1 amplification and sequencing have been described elsewhere (Apakupakul et al. Citation1999; Borda and Siddall Citation2004). Sequences of 42 species of Helobdella from previous studies were also included in the present analyses for comparative purposes. COX1 sequences of Haementeria ghilianii de Filippi, 1849 and Haementeria gracilis Cordero, 1941 were used to root the analysis since they constitute the sister group of Helobdella (Siddall and Borda Citation2003; Siddall et al. Citation2005).
Alignment, neighbor-joining analysis, and recognition of molecular characteristic attributes
All of the COX1 sequences obtained for this study, as well as sequences obtained from GenBank, were aligned with MUSCLE (Edgar Citation2004) on the European Bioinformatics Institute webserver (http://www.ebi.ac.uk/Tools/muscle/index.html), applying default settings. A neighbor-joining tree was calculated in PAUP* (Swofford Citation2002) using the Kimura two-parameter model of nucleotide substitution (Kimura Citation1980) following previous barcoding studies (Hebert et al. Citation2004). All of the distance values among COX1 sequences were calculated in PAUP* using the Kimura two-parameter model. Diagnostic nucleotide positions for particular clusters (molecular synapomorphies) were determined through the implementation of the Characteristic Attribute Organization System software (Sarkar et al. Citation2002a,Citationb).
Results
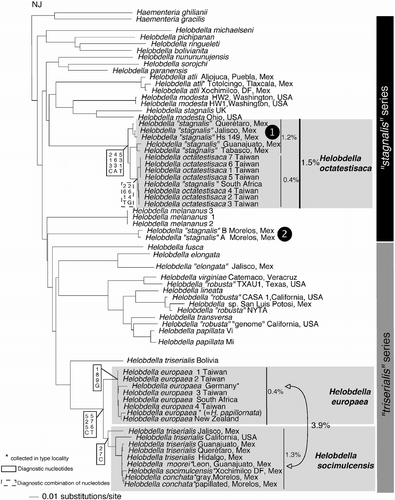
The neighbor-joining tree () resulting from the analyses of COX1 sequences of 63 samples of Helobdella rooted with two species of Haementeria recovers as a cluster all of the species of Helobdella. The “stagnalis” series was represented as a paraphyletic assemblage relative to a monophyletic “triserialis” series. Samples of Helobdella “stagnalis” from Mexico were found in two separate parts of the tree. The first forming a paraphyletic group (Helobdella “stagnalis 1”) presents 1.2% of genetic variation within its members and appears closely related to Helobdella octatestisaca from Taiwan and one sample from South Africa. The genetic variation within the latter group averaged 0.4%, and between those and H. “stagnalis 1” the genetic distance averaged 2.2%. The second cluster (Helobdella “stagnalis 2”) was found nestled between the “stagnalis” and “triserialis” series. Within the “triserialis” series, H. virginiae and Helobdella sp. from San Luis Potosi, Mexico, appeared grouped with various forms of Helobdella “robusta”, Helobdella lineata, Helobdella papillata, and Helobdella transversa from the USA. H. elongata from Mexico appeared in the same cluster with H. elongata from the USA, these two specimens having a genetic distance of 7%. H. moorei, H. conchata, H. socimulcensis and forms of H. triserialis from several localities of Mexico, and an unidentified leech from San Francisco, California grouped in a single cluster with an average of 1.3% genetic distance among them. This cluster appeared most closely related (average distance of 3.9%) to the genetically homogeneous ( < 0.4% within-group genetic distance) and globally invasive Helobdella europaea Kutschera, 1985.
Figure 1. Neighbor-joining tree based on the Kimura two-parameter substitution model of the COX1 locus of representative species of Helobdella showing the “stagnalis” and “triserialis” series. Numbers next to straight lines indicate average genetic distance within samples of a particular cluster. Number next to arrows indicates average genetic distance of pairwise comparisons between members of the two different clusters (H. europea and H. socimulcensis). Asterisks indicate specimens collected in the type locality. Numbers next to vertical lines indicate average genetic distances.
Discussion
The “stagnalis” series
There has historically been considerable taxonomic confusion surrounding the name H. stagnalis, and the fact that specimens identified as H. stagnalis fall into three different parts of the tree may partly reflect this confusion. This species was described by Linnaeus in 1758 based on common European specimens. Historically, the presence of a conspicuous chitinous scute on the dorsal surface, as seen on leeches collected in several areas of the world, would lead to diagnosis as H. stagnalis such that this has been considered a cosmopolitan species (Sawyer Citation1986). For example, a nearly indistinguishable leech described as Helobdella modesta Verrill, 1872 was later synonymized under H. stagnalis (Klemm Citation1972, Citation1982; Sawyer Citation1986). Siddall et al. (Citation2005) found a high degree of genetic variation between H. stagnalis collected from the UK relative to those from Ohio, USA and reestablished Verrill's name H. modesta for North American species (see also Madill and Hovingh Citation2007; CitationOceguera-Figueroa et al. in press). Mexican scutiferous samples were found in three different parts of the tree as putatively distinct species: H. atli, H. “stagnalis 1”, and as-yet undescribed species from Temixco, Morelos here designated as H. “stagnalis 2”. All three samples of H. atli, including that collected in Totolcingo, Tlaxcala, the type locality for the species, were found forming a single cluster next to the lineage that includes H. modesta and European H. stagnalis.
Helobdella “stagnalis” from several localities in Mexico clustered with the recently described H. octatestisaca Lai & Chang, 2009 from Taiwan, a lineage that also included a South African specimen. Lai et al. (Citation2009) suggested that H. octatestisaca might be a recently introduced species in Taiwan, because neither exhaustive fieldwork nor thorough examinations of scientific collections had previously uncovered this species. The extremely low genetic variation (0.4%) within the samples of H. octatestisaca and the sample from South Africa contrasts with the 1.5% among the whole cluster when also including the Mexican samples. This fact is in agreement with previous studies in a variety of organisms, including H. europaea (Siddall and Budinoff Citation2005; see ), predicting that invasive species show relatively low genetic variation compared with that of their source population (Tsutsui et al. Citation2000; Suarez and Tsutsui Citation2008).
The “triserialis” series
Helobdella triserialis was originally described based on specimens collected in Chile. However, because of the high degree of pigment variation throughout its presumed range, Ringuelet (Citation1943) recognized at least four subspecies. Siddall and Borda (Citation2003) found that North and South American forms constitute distinct evolutionary lineages and expanded Verrill's (1872) name Helobdella papillata for North American representatives. Surprisingly, H. elongata is included in this cluster notwithstanding its unusual morphology and is only distantly related (>2.5% genetic distance) to the morphologically similar (and formerly congeneric under Gloiobdella) South American counterpart Helobdella michaelseni. This suggests that several morphological attributes (i.e. cylindrical body, unpigmented teguments, and absence of gastric ceca) are unreliable indicators of recent diversification.
H. robusta is perhaps the best-known lophotrochozoan model organism. Efforts to understand the complex developmental mechanisms of this species culminated with the sequencing of its full genome (Weisblat and Kuo Citation2009). Bely and Weisblat (Citation2006) have demonstrated that at least three different lineages of leeches previously considered to be H. robusta have been independently employed in developmental biology research. Complicating this issue was that two distinct and unrelated COX1 lineages of H. robusta are found in the type locality in Sacramento, CA, USA (“CASA 1” and “genome” in ). In the absence of a more detailed morphological analysis of each of the different lineages in comparison with the holotype, the problem of which lineage is the real H. robusta remains unresolved. Indeed, the full genome that was sequenced may well belong to an undescribed species. H. virginiae and Helobdella sp. from San Luis Potosí, Mexico also appeared to be closely related to specimens of H. “robusta” from Texas and New York, respectively. In both cases, branch lengths suggest that they might represent independent species. Rather than quickly multiplying the number of species representing H. robusta on the basis of a single locus, we should also consider the possibility that H. virginiae, H. robusta, H. lineata, H. transversa, and H. papillata are capable of limited introgression to the extent that COX1 may not provide a reliable indication of species groups for this particular cluster.
In Mexico, three species morphologically similar to H. triserialis have been described. H. socimulcensis Caballero, 1931 from Xochimilco, D. F. and H. moorei Caballero, 1933 from León, Guanajuato, each were considered to be junior synonyms of Helobdella triserialis lineata by Ringuelet (Citation1981). The third species in this series is H. conchata Caballero, 1941 from Cuautla, Morelos, Mexico. Our results, including several samples for each name and including samples from the respective type localities, failed to recognize significant differences among them and strongly suggest that this entire group should be considered a single species. This cluster forms a lineage independent of H. triserialis sensu stricto (Bolivia) and, given the lack of morphological differences, the name H. socimulcensis Caballero, 1931 would be used for this group, which appears to be closely related to H. europaea.
Invasive species
The pattern of an invasive species with low genetic variation next to samples collected in their inferred natural habitat displaying high levels of genetic variation was found in two independent parts of the tree. H. octatestisaca and H. europaea were originally described from Taiwan and Germany, respectively. Both are geographic areas well removed from what appears to be their otherwise Helobdella robusta New World distribution. In both cases, Mexican samples appeared next to the putative invasive species clusters, but in any case identical COX1 sequences were found across them. Even though the general pattern in both parts of the tree seems similar, a closer analysis of each case would give different results. In both cases, the genetic distance between the Mexican populations and the invasive species averages more than 2%, a number that seems high enough to suggest the presence of multiple species (Hebert et al. Citation2004). Furthermore, in both cases, each group taken as a whole presents diagnostic nucleotides; in the case of the H. octatestisaca cluster, position 213 of the alignment presents a cytosine (C), 463 an adenine (A), and 531 a thymine (T), while the cluster H. europaea+H. socimulcensis presents two diagnostic nucleotides at positions 525 (C) and 576 (T). The difference between the two cases is that the cluster of H. europaea has an exclusive guanine (G) at position 189, whereas H. “stagnalis 1” samples form a paraphyletic assemblage. In addition, H. socimulcensis presents a diagnostic C at position 27, but on the contrary H. octatestisaca lacks a diagnostic nucleotide, but exhibits a diagnostic combination of T and G at positions 261 and 264. With this collective information in mind, it seems reasonable to suggest the renaming of the Mexican samples of H. “stagnalis 1” as H. octatestisaca, but keeping different names for the invasive species H. europaea and for the Mexican samples that, in agreement with their genetic similarity, should be renamed H. socimulcensis.
The use of DNA barcoding to identify species relies on the assumption that COX1 variation between species (i.e. interspecific) exceeds by a considerable amount the variation present within species (i.e. intraspecific). Although the straight use of genetic distances (>2%) as a criterion to differentiate species would lead to considering H. “stagnalis 1” as a species independent of H. octatestisaca, we think this could be an overestimation of species-level biodiversity. Using a discrete, fixed character-based approach represents, in our opinion, a better option, because it is in agreement both with the philosophical approaches of modern methods of phylogenetic analyses and with the need for diagnosis in classical taxonomy.
In conclusion, at least seven species of Helobdella occur in Mexico: H. atli, H. octatestisaca, H. virginiae, H. elongata, H. socimulcensis, and two forms diagnosed only with molecular data—Helobdella sp. from San Luis Potosi and H. “stagnalis” from Temixco, Morelos.
Acknowledgements
Patricia Escalante, Sergios-Orestis Kolokotronis, and Rob DeSalle kindly invited the authors to participate in this special issue. Sebastian Kvist and Anna Phillips contributed with their comments that greatly improved the quality of the paper. Florencia Bertoni, Elisa Cabrera, Gerardo Rivas, Rogelio Rosas, Salvador Díaz Cuevas, Jean Paul, and Giovanni Rosas collected specimens included in the present study. Luis García Prieto from the Colección Nacional de Helmintos, UNAM, Mexico assisted with the catalog numbers and collection management.
Declarations of interest: The present project was supported by the National Science Foundation (DEB grant numbers 01613802 and 0640436). A.O.-F. thanks CONACyT, Mexico (grant number 172322) and the CUNY Graduate Center Science Fellowship Program for financial support.
References
- Apakupakul K, Siddall ME, Burreson EM. 1999. Higher level relationships of leeches (Annelida: Clitellata: Euhirudinea) based on morphology and gene sequences. Mol Phylogenet Evol. 12:350–359.
- Bely AE, Weisblat DA. 2006. Lessons from leeches: A call for DNA barcoding in the lab. Evol Dev. 8:491–501.
- Borda E, Siddall ME. 2004. Arhynchobdellida (Annelida: Oligochaeta: Hirudinida): Phylogenetic relationships and evolution. Mol Phylogenet Evol. 30:213–225.
- Edgar RC. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 32:1792–1797.
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 101:14812–14817.
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Klemm DJ. 1972. Freshwater leeches (Annelida: Hirudinea) of North America . In :Biota of freshwater ecosystems, identification manual 8. Washington, DC: Environmental Protection Agency. p 53.
- Klemm DJ. 1982. The leeches (Annelida: Hirudinea) of North America. Cincinnati, OH: 172 pp Aquatic Biology Section, Environmental Monitoring and Support Laboratory, Office of Research and Development, US Environmental Protection Agency.
- Lai YT, Chang CH, Chen JH. 2009. Two new species of Helobdella Blanchard 1896 (Hirudinida: Rhynchobdellida: Glossiphoniidae) from Taiwan, with a checklist of hirudinea fauna of the island. Zootaxa. 2068:27–46.
- Madill J, Hovingh P. 2007. Freshwater leech (Annelida: Hirudinida) distribution in the Canadian Province of Newfoundland and Labrador and adjacent regions: Check-list, new records, new pigmentation forms, and Pleistocene refugia. Zootaxa. 1657:1–21.
- Oceguera-Figueroa A. 2007. Especie nueva del género Helobdella (Rhynchobdellida: Glossiphoniidae) del lago de Catemaco, Veracruz, México. Acta Zool Mex (nueva serie). 23:15–22.
- Oceguera-Figueroa A, León-Règagnon V. 2005. A new freshwater leech species of Helobdella (Annelida: Glossiphoniidae) from central Mexico. Zootaxa. 976:1–8.
- Oceguera-Figueroa A, Kvist S, Watson S, Sankar DF, Overstreet RM, Siddall ME. Leech collections from Washington State, USA with the description of two new species of Placobdella (Annelida: Glossiphoniidae). Am Mus Novit. in press In press.
- Ringuelet RA. 1943. Sobre la morfología y variabilidad de Helobdella triserialis (Em. Bl) (Hirudinea, Glossiphoniidae). Notas Mus Plata. 8:215–240.
- Ringuelet RA. 1981. Clave para el reconocimiento de los hirudíneos de México. An Inst Biol Univ Nac Autón Méx Ser Zool. 52:89–97.
- Ringuelet RA. 1985. Fauna de agua dulce de la República de Argentina, Hirudinea. Buenos Aires: FECIC. p 321.
- Sarkar IN, Planet PJ, Bael TE, Stanley SE, Siddall M, DeSalle R, Figurski DH. Characteristic attributes in cancer microarrays. J Biomed Inform. 2002a; 35:111–122.
- Sarkar IN, Thornton JW, Planet PJ, Figurski DH, Schierwater B, DeSalle R. An automated phylogenetic key for classifying homeoboxes. Mol Phylogenet Evol. 2002b; 24:388–399.
- Sawyer RT. 1986. Leech biology and behavior. Oxford: Clarendon Press. p 1065.
- Siddall ME, Borda E. 2003. Phylogeny and revision of the leech genus Helobdella (Glossiphoniidae) based on mitochondrial gene sequences and morphological data and a special consideration of the triserialis complex. Zool Scr. 32:23–33.
- Siddall ME, Budinoff RB. 2005. DNA-barcoding evidence for widespread introductions of a leech from the South American Helobdella triserialis complex. Conserv Genet. 6:467–472.
- Siddall ME, Budinoff RB, Borda E. 2005. Phylogenetic evaluation of the systematics and biogeography of the leech family Glossiphoniidae. Invert Syst. 19:105–112.
- Suarez AV, Tsutsui ND. 2008. The evolutionary consequences of biological invasions. Mol Ecol. 17:351–360.
- Swofford DL. 2002. PAUP* phylogenetic analysis using parsimony (*and other methods), Ver. 4.0b10. Sunderland, MA: Sinauer Associates.
- Tsutsui ND, Suarez AV, Holway DA, Case TJ. 2000. Reduced genetic variation and the success of an invasive species. Proc Natl Acad Sci USA. 97:5948–5953.
- Weisblat DA, Kuo DH. Helobdella (leech): A model for developmental studies. Cold Spring Harb Protoc. 2009 pdb.emo121.
