Abstract
Background and aims: Considering the promising use of DNA barcoding for species identification, the importance of the freshwater fish fauna of the Paraíba do Sul River Basin, and its advanced stage of degradation, the present study evaluated the effectiveness of DNA barcoding to identify the fish species in this basin.
Materials and methods: A total of 295 specimens representing 58 species belonging to 40 genera, 17 families, and 5 orders were sequenced.
Results: The DNA barcodes discriminated all species analyzed without ambiguity. The results showed a pronounced difference between conspecific and congeneric pair-wise sequence comparisons, demonstrating the existence of a “barcode gap” for the species analyzed. The nearest-neighbor distance analysis showed only three cases with Kimura two-parameter values lower than a 2% divergence threshold. However, the patterns of divergence observed in each case remained sufficient to discriminate each species, revealing the accuracy of DNA barcoding even cases with relatively low genetic divergence. At the other extreme, three species displayed high genetic sequence divergence among conspecifics. For two cases, Characidium alipioi and Geophagus proximus, barcoding proved effective at flagging possible new species. For another case, Astyanax bimaculatus, the use of DNA barcoding of the comparison of shared freshwater fish fauna between different basins revealed itself as highly useful in disclosing that the previously identified A. bimaculatus “cluster A” probably represents the species Astyanax altiparanae.
Conclusion: The present study is among the first to assess the efficiency of barcoding for the Brazilian freshwater fishes. The results demonstrate the utility of barcoding to identify the fauna from this basin, contribute to an enhanced understanding of the differentiation among species, and to help flag the presence of overlooked species.
Introduction
Species are the fundamental unit of comparison in biology, from anatomy to behavior, development, ecology, evolution, genetics, molecular biology, paleontology, physiology, systematic, and so forth (De Queiroz Citation2005). Thus, the capability to correctly identify species is crucial in order to minimize “error cascades” resulting from the use of bad taxonomy in science (e.g. Bortolus Citation2008). Specifically, such errors are known to have a significant impact on the population assessment of overfished species (e.g. Beerkircher et al. Citation2009).
Traditionally, for the identification of species, morphological characters are used. However, the development of molecular biology created a new set of useful tools to identifying species.. Protein electrophoresis on starch gel was first used more than 45 years ago to identify species (Manwell and Baker Citation1963). Since then, many studies have been published using a diverse assemblage of molecular approaches and markers to identify species, such as allozymes (Aron and Sole-Cava Citation1991; Gusmão et al. Citation2000), restriction fragment length polymorphism (Moysés and Almeida-Toledo Citation2002), DNA arrays (Hajibabei et al. Citation2007), single nucleotide polymorphism (Shaffer and Thonsom Citation2007), multiplex PCR (Mendonça et al. Citation2009), DNA sequences (Pook and McEwing Citation2005; Lemer et al. Citation2007), and many others.
Given this promising but heterogeneous scenario, Hebert et al. (Citation2003) proposed the use of a small fragment (∼648 bp) from the 5′ end of the mitochondrial cytochrome c oxidase subunit I (COI) gene as a universal standard to identify most species of animals. Because these sequences tend to vary between species but are relatively invariant within species, a reference sequence library derived from expert-identified reference material can be used to infer the identity of an unknown specimen by matching its sequence to the reference library, hence the use of the “barcode” metaphor. The barcode of life data system (BOLD; Ratnasingham and Hebert Citation2007) is a collaborative online workbench for the assembly and curation of a global barcode sequence library that aims to facilitate and automate the process of species identification. The Fish Barcode of Life (FISH-BOL) campaign (Ward et al. Citation2009) advocates the use of both COI and BOLD by the ichthyological community for constructing a global reference sequence library designed to enable the rapid, accurate, and cost-effective identification of eggs, larval, juvenile, and fragmentary remains, even by non-specialists. Many studies have proven the effectiveness of barcoding to identify fishes (Ward et al. Citation2005, Citation2009; Hubert et al. Citation2008; Valdez-Moreno et al. Citation2009), yet much work remains to be done.
Brazil has a mega freshwater fish fauna with about 2587 described species and many others yet to be described (Buckup et al. Citation2007). Many researchers suggest that this number will increase in the future (Schaefer Citation1998; Vari and Malabarba Citation1998; Castro et al. Citation2003, Citation2004, Citation2005; Langeani et al. Citation2007), since the sampling of species is insufficient and many regions remain almost unexplored (Junk Citation2007; Langeani et al. Citation2007). The Paraíba do Sul River Basin is located at latitudes between 20°26′ and 23°39′ S and longitudes between 41°00′ and 46°30′ W in southeastern Brazil (Teixeira et al. Citation2005). This basin drains an area of about 57,000 km2 and has about 1200 km of extension up to its mouth in the Atlantic Ocean and is located in one of the most urbanized and industrialized areas of Brazil with serious anthropogenic impacts. The most recent survey of freshwater fish fauna showed 81 species in this basin belonging to 55 genera, 29 families, and 9 orders (Teixeira et al. Citation2005). Many of these species are economically important, many exhibit migratory behavior, and some are at risk of extinction (Bizerril and Primo Citation2001). Considering the promising use of DNA barcoding to identify species and the importance of the freshwater fish fauna of the Paraíba do Sul River Basin and its advanced stage of degradation, the present study aims to assemble a comprehensive reference sequence library for regional fishes and to evaluate its effectiveness for species identification.
Materials and methods
Specimen collection
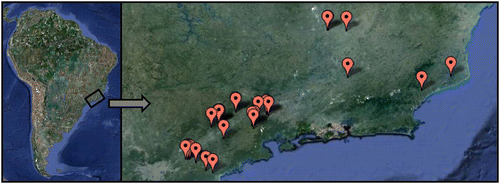
Some 295 fishes were collected from 27 sites in the Paraíba do Sul River Basin (). All specimens had a fresh tissue fragment removed and preserved in absolute ethanol at − 20°C. Morphological vouchers were deposited in the fish collection of Laboratório de Biologia e Genética de Peixes (LBP), Departamento de Morfologia, Instituto de Biociências, UNESP, Botucatu, São Paulo, Brazil. All specimens were identified with the help of taxonomists and identification keys. All procedures were performed according to local ethics committee. Specimen provenance data, including geospatial coordinates of collection sites and other relevant details, are recorded in the BOLD project “Fishes from Paraíba do Sul River, Brazil” (project code: FPSR).
Extraction, PCR amplification, and DNA sequencing
The DNA barcoding was carried out at the Canadian Centre for DNA Barcoding (CCDB), Canada, using the standard protocols (Hajibabaei et al. Citation2005) and at the LBP, UNESP, Botucatu, Brazil. Total genomic DNA was isolated from fin or muscle tissue of each specimen using one of two different methods: DNeasy Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions (LBP), and with vertebrate lysis buffer with proteinase K digested overnight at 56°C and subsequently extracted using a membrane-based approach on a Biomek NX Laboratory Automation Workstation using AcroPrep96 (Beckman Coulter, Inc., Brea, California, USA), and 1-ml filter plates with a 10-mm PALL glass fiber media (Ivanova et al. Citation2006; CCDB). A portion (648 bp) of the 5′ end of the mitochondrial COI gene was amplified by PCR using different combinations of fish primers: FishF1, FishR1, FishF2, FishR2 (Ward et al. Citation2005) or the M13-tailed primer cock-tails C_FishF1t1–C_FishR1t1 and C_VF1LFt1–C_VR1LRt1 (Ivanova et al. Citation2007). The 12.5 μl PCR mixes include 6.25 μl of 10% trehalose, 2 μl ultrapure water, 1.25 μl of 10 × PCR buffer, 0.625 μl MgCl2 (50 mM), 0.125 μl each primer (0.01 mM), 0.0625 μl each dNTP (0.05 mM), 0.625 μl Taq polymerase, and 2.0 μl DNA template. PCR was carried out in a thermocycler (Veriti 96-Well Thermal Cycler; Applied Biosystems, Inc., Foster City, California, USA). The PCR conditions followed the Hajibabaei et al. (Citation2005) protocols. Amplified products were checked on 1% agarose gels. In the LBP, the PCR products were purified with ExoSap-IT (USB Corp., Cleveland, OH, USA) following the manufacturer's protocol. At the CCDB, PCR products were labeled with the BigDye Terminator v3.1 Cycle Sequencing Ready Reaction kit (ABI) using standard methods (Hajibabaei et al. Citation2005), and were bidirectionally sequenced on an ABI 3730 DNA Analyzer. At the LBP, the cycle sequencing reaction was carried out using a BigDye Terminator v3.1 Cycle Sequencing Ready Reaction kit (ABI) in a final volume of 7 μl containing 1.4 μl template, 0.35 μl primer (10 μM), 1.05 μl buffer 5 × , 0.7 μl BigDye mix, and water. The cycle sequencing conditions included initial denaturation at 96°C for 2 min followed by 35 cycles of denaturation at 96°C for 45 s, annealing at 50°C for 60 s, and extension at 60°C for 4 min. The PCR sequencing products were purified with ethylenediamine tetraacetic acid/sodium acetate/alcohol following the protocol suggested in the BigDye Terminator v3.1 Cycle Sequencing kit's manual. All samples were sequenced on an ABI 3130 Genetic Analyzer capillary sequencer following the manufacturer's instructions. Sequence data, trace files, primer details, photographs, and collection localities for specimens are available within the project FPSR on BOLD (www.barcodinglife.org). Sequences have also been deposited on GenBank (accession numbers: GU702075–GU702317, HM064974–HM065026).
Data analysis
All sequences were analyzed using SeqScape software v2.6 (ABI) to obtain the consensus sequences and check the occurrence of deletions, insertions, and stop codons. The sequences were aligned using the tools available on the BOLD v2.5 web site (http://www.barcodinglife.org). The genetic distances among and within species were calculated using the Kimura two-parameter (K2P) substitution model (Kimura Citation1980). A neighbor-joining (NJ) tree of K2P distances was created to provide a graphic representation of the patterning of divergence between species with the software MEGA v4.0 (Tamura et al. Citation2007).
Results
A total of 58 species belonging to 40 genera, 17 families, and 5 orders were sampled and barcoded (Appendices 1 and 2; ; FPSR project on BOLD). Fifty-two species were represented by more than one specimen (average ∼5 specimens/species) reaching a total of 295 COI sequences (∼652 bp; Appendices 1 and 2; ; FPSR project on BOLD). The mitochondrial COI sequences obtained were of high quality and the subsequent contig assemblies generated using them did not exhibit any evidence of insertions, deletions, or stop codons.
Figure 2. Compact K2P distance NJ tree showing the 58 analyzed species from the Paraíba do Sul River Basin. In parentheses are the numbers of specimens sequenced. *Species also reported in the Upper Paraná River Basin.
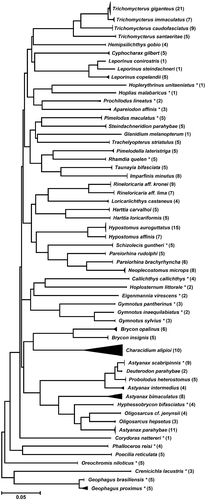
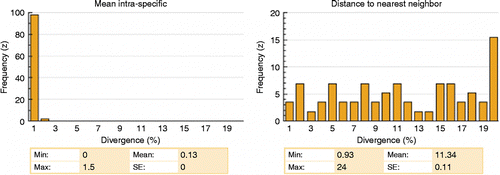
All species were discriminated by the barcode sequences. A compact NJ tree is presented in and the whole NJ tree is supplied in Appendix 2. The average K2P distance of specimens within species was 0.13% compared with 10.36% for species within genera (). The overall variation was about 79 times greater among congeneric species than within species (). A steady increase in genetic variation through increasing taxonomic levels was observed, supporting a marked change of genetic divergence at the species boundaries (). The nearest-neighbor distance analysis (NND), namely the minimum genetic distance between a species and its closest neighbor species, was carried out to analyze the distribution of genetic divergence. The analysis showed only three cases with K2P genetic divergence lower than 2% (), but enough to separate these species. The NND analysis also showed that 98% of conspecific comparisons were lower than 1% genetic distance ().
Table I. K2P genetic divergence values within different taxonomic levels from 295 specimens of Paraíba do Sul River Basin analyzed.
Three species displayed high K2P genetic divergence within specimens that had been assigned to a single species ( and ). Characidium alipioi displayed two subclusters () with high K2P divergence between them (10.2%) compared with < 0.1% within each cluster (). In another case, Geophagus proximus also displayed two subclusters () with 1.1% average K2P divergence between them compared with < 0.1% within each cluster (). Although these values are relatively low, they are much higher than inter-cluster values and very similar with average K2P divergence with Geophagus brasiliensis, its congeneric species (1.7%; ). These two cases represent clearly two possible new species to Paraíba do Sul river Basin as revealed by DNA barcode technique.
Figure 4. K2P distance NJ tree of species with high conspecific genetic divergence. Voucher numbers indicated before species name. Astyanax altiparanae from the Upper Paraná Riber Basin.
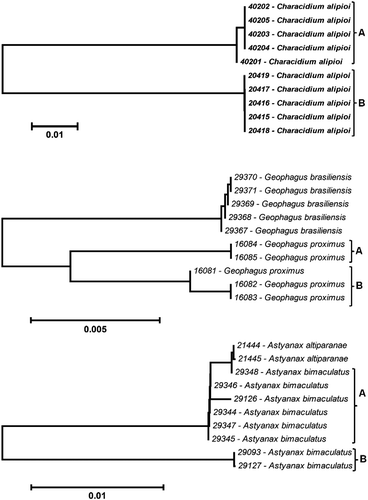
Table II. K2P genetic divergence between subclusters obtained for C. alipioi, G. proximus and A. bimaculatus.
In the third case, Astyanax bimaculatus displayed two subclusters () with an average K2P genetic divergence of 3.7% between them, while the average K2P genetic divergence within each subcluster was < 0.1% (). Additionally, we compared the A. bimaculatus COI sequences with two COI sequences of Astyanax altiparanae specimens from the Upper Paraná River Basin (neighboring basin) to check the similarity among these pairs of species. The analysis showed that A. bimaculatus subcluster A displayed high similarity with A. altiparanae (K2P divergence = 0.2%; and ), indicating that these two sets of samples might belong to the same species.
We also compared the G. proximus and C. alipioi species with specimens belonging to the Upper Paraná River Basin (neighboring basin), but they showed no similarity with these specimens (data not shown) – reinforcing the hypothesis of possible new species for these cases.
Discussion
We assessed 58 out of 81 species (70.6%) ascribed (Teixeira et al. Citation2005) to the Paraíba do Sul River Basin. The DNA barcode sequences were very effective, correctly discriminating all 58 species analyzed (). The average K2P divergence within conspecific specimens was only 0.13%, a value 79 times lower than that found among congeneric species (10.36%; ). These values are consistent with those found in the literature. Ward et al. (Citation2005) found average values of 0.39 and 9.93% by conspecific and congeneric comparisons, respectively, for marine fishes, discriminating all species. Hubert et al. (Citation2008), studying freshwater fishes from Canada, found average K2P values of 0.27% (conspecific) and 8.37% (congeneric), discriminating 93% of the species. In another study, Valdez-Moreno et al. (Citation2009) assessed the freshwater fishes from Mexico and Guatemala and found average K2P values of 0.45% (conspecific) and 5.1% (congeneric), discriminating 93% of the species. More recently, Lara et al. (Citation2010) assessed the freshwater fishes from Cuba and found average K2P values of 0.6% (conspecific) and 9.1% (congeneric), discriminating 96% of the species, while Ward et al. (Citation2009) analyzed the values of K2P genetic divergence of 1088 fish species available on BOLD and found average K2P values of 0.3% (conspecific) and 8.4% (congeneric), discriminating about 97.5% of the species. These values are very similar with those found in the present study, showing that such values may be a pattern for fish species. The difference between conspecific and congeneric average K2P divergence was more pronounced in our analysis (79-fold difference) compared with values found in the other fish studies (Ward et al. Citation2005; Hubert et al. Citation2008; Valdez-Moreno et al. Citation2009; Ward Citation2009). This fact is attributable to a low average K2P distance value found in the conspecific comparison (only 0.13%). This relatively low value may be ascribed to the possibility that the species analyzed here have a low conspecific variation or because of a limited sampling of the genetic variation for many species. Although we had tried to sample the maximum variation within each species, with most of species represented by more than one specimen (average ∼5 specimens/species), it is possible that an increase in the number of specimens and geographical areas sampled led to an increase in conspecific values.
Our results clearly showed the existence of a barcode gap, facilitating the unambiguous discrimination of the species analyzed (). The barcode gap is confirmed by the NND analysis with an average K2P distance to nearest neighbor of 11.34 (ranged from 0.93 to 24%), while 98% of the conspecific comparison showed values lower than 1% (). The NND analysis showed three cases with K2P genetic distance lower than 2% (). The Trichomycterus giganteus and Trichomycterus immaculatus species showed only 0.93% of K2P divergence between them. The Astyanax scabripinnis and Deuterodon parahybae species and G. brasiliensis and G. proximus species displayed K2P divergence of 1.4 and 1.56%, respectively. Although low, these values allowed the discrimination of these pairs of species, which showed high differences between conspecific and congeneric K2P divergence values (conspecific values < 0.1%). Ward et al. (Citation2009) showed that about 17% of the genetic divergence values among congeners are lower than 3% and 3.7% of the congeners' comparisons, which are lower than 1%, consistent with the three cases reported here. Krieger and Fuerst (Citation2002) observed in their work that different teleost orders have different evolutionary rates and that it is possible that different families, genera, and species could have different evolutionary rates, which could explain the low values found in some pairs of species. Moreover, these low values may indicate a recent radiation for these species, as reflected by the low K2P divergences observed between pairs of species. Montoya-Burgos (Citation2003) and Hubert et al. (Citation2007) pointed to a recent radiation of some groups of freshwater fishes in South America and suggested that these patterns could be valid for most Neotropical freshwater fishes. Perdices et al. (Citation2002, Citation2005) and Ornelas-Gacia et al. (Citation2008) proposed similar patterns for Mesoamerica.
Three species displayed high conspecific K2P genetic divergence ( and ). Hebert et al. (Citation2004) suggested a threshold to delimit species with DNA barcode data. These values should be at least 10 times the average conspecific values. Ward et al. (Citation2009) analyzed the genetic divergence distribution among “barcoded” fishes and suggested that if the unknown specimen is more than 2% divergent from a known specimen, it is very likely that this is a different species with a probability greater than 95%. The two subclusters of C. alipioi () displayed very high average K2P divergence between them (10.2%) compared with < 0.1% within each cluster (), flagging clearly a possible new species. The second species that showed high conspecific K2P genetic distance was G. proximus, which also displayed two subclusters () with 1.1% average K2P divergence between them compared with < 0.1% within each cluster (). This subcluster divergence value is relatively low, but still larger than observed intracluster values and is in fact very similar with the average K2P divergence with its congeneric species, G. brasiliensis (1.7%; ), also flagging a possible new species. These species do not meet the 2% heuristic thresholds of CitationHebert et al. (2004) and/or CitationWard et al. (2009); however, we believe that, in this case, the most important observation is in regard to the congeneric K2P value comparisons, which are very similar with those found between the two subclusters of G. proximus (). Thus, standard threshold values must be applied carefully and other characteristics must also be considered. Ornelas-Garcia et al. (Citation2008), working with species of the genus Astyanax from Mesoamerica, found that some specimens formed separate clusters and suggested the occurrence of a species complex in this genus, assigning provisional names to each cluster obtained. Ward et al. (Citation2008), working with Asian sea bass Lates calcarifer specimens from different localities (Australia and Myanmar), found genetic distance values of 9.5% between two groups for COI (DNA barcode region) and 11.3% for cytochrome b. The authors suggested the existence of two species. Many species were discovered with the use of molecular data and some were formally described later (Smith et al. Citation2005; Witt et al. Citation2006; Kon et al. Citation2007; Ward Citation2007; Nguyen and Seifert Citation2008; Ward et al. Citation2008; Yassin et al. Citation2008). The DNA barcode was also utilized as part of the validation and formal description of new fish species such as Coryphopterus kuna (Victor Citation2007); Urolophus kapalensis (Yearsley and Last Citation2006); Brachionichthys autralis (Last et al. Citation2007); five new species of Chromis genus (Pyle et al. Citation2008), Dipturus argentinensis (Diaz de Astarloa et al. Citation2008) and Moenkhausia forestii (Benine et al. Citation2009). Thus, our results showed clearly the usefulness of DNA barcoding as a technique for the flagging of possible new species contributing to our knowledge of this basin's fish biodiversity.
On the other hand, the third case that displayed high conspecifc K2P divergence was A. bimaculatus (). The two subclusters obtained differ by 3.7% from each other compared with < 0.1% within each subcluster (). This value is consistent with Hebert et al.'s (Citation2004) and Ward et al.'s (Citation2009) suggestion for flagging possible new species. However, the comparison of A. bimaculatus subclusters with A. altiparanae specimens from the Upper Paraná Basin (neighboring basin; ) showed that A. bimaculatus “subcluster A” displayed high similarity with A. altiparanae (K2P divergence = 0.2%; ). This result suggests that the A. bimaculatus cluster A is probably the A. altiparanae species. Ab'Saber (Citation1957) discusses the hypothesis that, in the past, the Paraíba do Sul River Basin and the Upper Paraná River Basin (in its Upper Tietê River portion) were connected. Moreover, very recent connections during the Quaternary between rivers in the Upper Tietê River area were suggested (Ribeiro et al. Citation2006). Additionally, we checked the FishBase online database (http://www.fishbase.org) and other studies concerning the composition of the ichthyofauna of Brazil, mainly the Upper Paraná Basin (Reis et al. Citation2003; Nelson Citation2006; Langeani et al. Citation2007) and found at least 21 shared species between the Upper Paraná River and Paraíba do Sul River Basins ( and Appendix 1). These facts reinforce the hypothesis that the A. bimaculatus “cluster A” actually represents the A. altiparanae species. However, we must consider the hypothesis that some of these shared species represent different species with the same name. In those cases, a further detailed comparison between specimens from both basins will be necessary. An alternative explanation to the presence of the A. altiparanae in the Paraíba do Sul River would be the introduction of species, which should also be further tested. Our results show a useful and promising application of the DNA barcoding technique to the Brazilian ichthyofauna through the comparison of shared fish fauna in the Brazilian hydrographic basins. This approach will contribute to an enhanced understanding of the geographical distribution of many species and will also help flag potentially overlooked species.
Conclusions
The present study is the first to assess the effectiveness of DNA barcoding for the identification of the mega Brazilian freshwater fish fauna following the procedural recommendations of FISH-BOL to meet community standards developed for barcoding (CitationHanner 2005). This stands in contrast to other recent studies (e.g. Ardura et al. Citation2010) where results are reported, but data are not deposited in publicly accessible databases (despite claims to the contrary). Our results evidenced the usefulness of DNA barcodes for cataloging Brazilian freshwater fish species and for identifying those groups that deserve further taxonomic attention. Significantly, they also democratize access to biological identifications by the broader conservation and management community, which is predicted to result in broad socio-economic impacts (Costa and Carvalho Citation2007).
Supplementary Material
Download MS Word (446.5 KB)Supplementary Material
Download PDF (105.1 KB)Acknowledgements
The authors are grateful to Ricardo Benine and Osvaldo T. Oyakawa for their help with the fish identification. They would like to thank the CCDB team, especially Constantine Christopoulos, for collaboration with the acquisition of many sequences. Financial support for the present study was provided by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); and by funding to the Canadian Barcode of Life Network from the Natural Sciences and Engineering Research Council of Canada (NSERC) and other sponsors listed at www.BOLNET.ca.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ab'Saber AN. 1957. O problema das conexões antigas e da separação da drenagem do Paraíba e Tietê. Bol Paul Geogr. 26:38–49.
- Ardura A, Linde AR, Moreira JC, Garcia-Vazquez E. 2010. DNA barcoding for conservation and management of Amazonian commercial fish. Biol Conserv. 143:1438–1443.
- Aron SL, Solé-Cava AM. 1991. Genetic evaluation of the taxonomic status of two varieties of the cosmopolitan ascidian Botryllus niger (Ascidiacea: Botryllidae). Biochem Syst Ecol. 19:271–276.
- Beerkircher L, Arocha F, Barse A, Prince E, Restrepo V, Serafy J, Shivji M. 2009. Effects of species misidentification on population assessment of overfished white marlin Tetrapturus albidus and roundscale spearfish T. Georgii. Endanger Species Res. 9:81–90.
- Benine RC, Mariguela TC, Oliveira C. 2009. Nem species of Moenkhausia Eigenmann, 1903 (Characiformes: Characidae) with comments on the Moenkhausia oligolepis species complex. Neotrop Ichthyol. 7 2: 161–168.
- Bizerril CRSF, Primo PBS. 2001. Peixes de águas interiores do estado do Rio de Janeiro. Rio de Janeiro: Fundação de Estudos do Mar417.
- Bortolus A. 2008. Error cascades in the biological sciences: The unwanted consequences of using bad taxonomy in ecology. Ambio. 37:114–118.
- Buckup PA, Menzes NA, Ghazzi MS. 2007. Catálogo das espécies de peixes de água doce do Brasil. Rio de Janeiro: Museu Nacional195.
- Castro RMC, Casatti L, Santos HF, Ferreira KM, Ribeiro AC, Benine RC, Dardis GZP, Melo ALA, Stopiglia R, Abreu TX, Bockmann FA, Carvalho M, Gibran FZ, Lima FCT. 2003. Estrutura e composição da ictiofauna de riachos do Rio Paranapanema, sudeste e sul do Brasil. Biota Neotrop. 3 1: 1–31 [Online]. Available at: http://www.biotaneotropica.org.br/v3n1/pt/abstract?article+BN0170301.
- Castro RMC, Casatti L, Santos HF, Melo ALA, Martins LSF, Ferreira KM, Gibran FZ, Benine RC, Carvalho M, Ribeiro AC, Abreu TX, Bockmann FA, Pelição GZ, Stopiglia R, Langeani F. 2004. Estrutura e composição da ictiofauna de riachos da bacia do Rio Grande, no Estado de São Paulo, Sudeste do Brasil. Biota Neotrop. 4 1: 1–39 [Online]. Available at7: http://www.biotaneotropica.org.br/v4n1/pt/abstract?article+BN0170402004.
- Castro RMC, Casatti L, Santos HF, Vari RP, Melo ALA, Martins LSF, Abreu TX, Benine RC, Gibran FZ, Ribeiro AC, Bockmann FA, Carvalho M, Pelição GZP, Ferreira KM, Stopiglia R, Akama A. 2005. Structure and composition of the stream ichthyofauna of four tributary rivers of the upper Rio Paraná basin, Brazil. Ichthyol Explor Freshw. 16:193–214.
- Costa FO, Carvalho GR. 2007. The barcode of life initiative: Synopsis and prospective societal impacts of DNA barcoding of fish. Genomics Soc Policy. 3:29–40.
- De Queiroz K. 2005. Ernst Mayr and the modern concept of species. Proc Natl Acad Sci USA. 102 s1: 6600–6607.
- Diaz de Astarloa JM, Mabragana E, Hanner R, Figueroa DE. 2008. Morphological and molecular evidence for a new species of longnose skate (Rajiformes: Rajidae: Dipturus) from Argentinean waters based on DNA barcoding. Zootaxa. 1921:35–46.
- Gusmão J, Lazoski C, Solé-Cava AM. 2000. A new species of Penaeus (Crustacea: Penaeidae) revealed by allozyme and cytochrome oxidase I analyses. Mar Biol. 137:435–446.
- Hajibabaei M, De Waard JR, Ivanova NV, Ratnasingham S, Dooh R, Kirk SL, Mackie PM, Hebert PDN. 2005. Critical factors for assembling a high volume of DNA barcodes. Philos Trans R Soc B Biol Sci. 360:1959–1967.
- Hajibabei M, Singer GAC, Clare EL, Hebert PDN. 2007. Design and applicability of DNA arrays and DNA barcodes in biodiversity monitoring. BMC Biol. 5:1–15.
- Hanner R. 2005. Proposed standards for BARCODE records in INSDC [Online]. Available at: http://www.barcoding.si.edu/PDF/DWG_data_standards-Final.pdf. Accessed on 17 July 2010.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc London B Biol Sci. 270:313–321.
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astrapes fulgerator. Proc Natl Acad Sci USA. 101:14812–14817.
- Hubert N, Duponchelle F, Nuñez J, Garcia-Davila C, Paugy D, Renno JF. 2007. Phylogeography of the piranha genera Serrasalmus and Pygocentrus: Implications for the diversification of the Neotropical ichthyofauna. Mol Ecol. 16:2115–2136.
- Hubert N, Hanner R, Holm E, Mandrak NE, Taylor E, Burridge M, Watkinson D, Dumont P, Curry A, Bentzen P, Zhang J, April J, Bernatchez L. 2008. Identifying Canadian freshwater fishes through DNA barcodes. PLoS ONE. 3:e2490.
- Ivanova NV, De Waard JR, Hebert PDN. 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes. 6:998–1002.
- Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. 2007. Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes. 7:544–548.
- Junk W. 2007. Freshwater fishes of South America: Their biodiversity, fisheries, and habitats – a synthesis. Aquat Ecosyst Manage. 10:228–242.
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Kon T, Yoshino T, Mukai T, Nishida M. 2007. DNA sequences identify numerous cryptic species of the vertebrate: A lesson from the gobioid fish Schindleria. Mol Phylogenet Evol. 44:53–62.
- Krieger J, Fuerst PA. 2002. Evidence for a slowed rate of molecular evolution in the order Acipenseriformes. Mol Biol Evol. 19:891–897.
- Langeani F, Castro RMC, Oyakawa OT, Shibatta OA, Pavanelli CS, Casatti L. 2007. Diversidade da fauna do Alto Rio Paraná: Composição atual e perspectivas futuras. Biota Neotrop. 7:181–197.
- Lara A, Ponce de Leon JL, Rodriguez R, Casane D, Cote G, Bernatchez L, Garcia-Machado E. 2010. DNA barcoding of Cuban freshwater fishes: Evidence for cryptic species and taxonomic conflicts. Mol Ecol Resour. 10:421–430.
- Last PR, Gledhill DC, Holmes BH. 2007. A new handfish, Brachionichthys australis sp. nov, (Lophiiformes: Brachionichthyidae), with a redescription of the critically endangered spotted handfish, B. hirsutus (Lacépède). Zootaxa. 1666:53–68.
- Lemer S, Aurelle D, Vigliola L, Durand JD, Borsa P. 2007. Cytochrome b barcoding, molecular systematics and geographic differentiation in rabbitfishes (siganidae). C R Biol. 330:86–94.
- Manwell C, Baker CMA. 1963. A sibling species of seacucumber discovered by starch–gel electrophoresis. Comp Biochem Physiol. 10:39–53.
- Mendonça FF, Hashimot DT, Porto-Foresti F, Oliveira C, Gadig OBF, Foresti F. 2009. Identification of the shark species Rizoprionodon lalandii and R. porosus (Elasmobranchii: Carcharinidae) by multiplex PCR and PCR–RFLP techniques. Mol Ecol Resour. 9:771–773.
- Montoya-Burgos JI. 2003. Historical biogeography of the catfish genus Hypostomus (Siluriformes: Loricariidae), with implications on the diversification of Neotropical ichthyofauna. Mol Ecol. 12:1855–1867.
- Moysés CB, Almeida-Toledo LF. 2002. Restriction fragment length polymorphisms of mitochondrial DNA among five freshwater fish species of the genus Astyanax (Pisces, Characidae). Genet Mol Biol. 25:402–407.
- Nelson JS. 2006. Fishes of the World. Hoboken, NJ: Wiley601.
- Nguyen HDT, Seifert KA. 2008. Description and DNA barcoding of three new species of Leohumicola from South Africa and the United States. Personia. 21:57–98.
- Ornelas-Garcia CP, Dominguez-Dominguez O, Doadrio I. 2008. Evolutionary history of the fish genus Astyanax Baird & Girard (1854) (Actynopterigii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evol Biol. 8:340.
- Perdices A, Bermingham E, Montilla A, Doadrio I. 2002. Evolutionary history of the genus Rhamdia (Teleostei: Pimelodidae) in Central America. Mol Phylogenet Evol. 25:172–189.
- Perdices A, Doadrio I, Bermingham E. 2005. Evolutionary history of the synbranchid eels (Teleostei: Synbranchidae) in Central America and the Caribbean islands inferred from their molecular phylogeny. Mol Phylogenet Evol. 37:460–473.
- Pook CE, McEwing R. 2005. Mitochondrial DNA sequences from dried snake venom: A DNA barcoding approach to the identification of venom samples. Toxicon. 46:711–715.
- Pyle RL, Earle JL, Greene BD. 2008. Five new species of the damselfish genus Chromis (Perciformes: Labroidei: Pomacentridae) from deep coral reefs in the tropical western Pacific. Zootaxa. 1671:3–31.
- Ratnasingham S, Hebert PDN. 2007. BOLD: The barcode of life data system. Mol Ecol Notes. 7:355–364 (www.barcodinglife.org).
- Reis RE, Ferraris C, Kullander SO. 2003. Check list of freshwater fishes of South and Central America. Porto Alegre: Edipucrs729.
- Ribeiro AC, Lima FCT, Riccomini C, Menezes NA. 2006. Fishes of the Atlantic Rainforest of Boracéia: Testimonies of the Quaternary fault reactivation within a Neoproterozoic tectonic province in Southeastern Brazil. Ichthyol Explor Freshw. 17:157–164.
- Schaefer SA. 1998. Conflict and resolution impact of new taxa on philogenetic studies of Neotropical cascudinhos (Siluriformes: Loricariidae). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS. editors. Phylogeny and Classification of Neotropical Fishes. Porto Alegre: Edipucrs375–400.
- Shaffer HB, Thonsom RC. 2007. Delimiting species in recent radiations. Syst Biol. 56:896–906.
- Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN. 2005. DNA barcodes reveal cryptic host-specificity within the presumed polyphagus members of a genus of parasitoid flies (Diptera: Tachinidae). Proc Natl Acad Sci USA. 103:3657–3662.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA 4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599.
- Teixeira TP, Pinto BCT, Terra BF, Estiliano EO, Gracia D, Araújo FG. 2005. Diversidade das assembléias de peixes nas quatro unidades geográficas do rio Paraíba do Sul. Iheringia Sér Zool. 95:347–357.
- Valdez-Moreno M, Ivanova NV, Elías-Guitiérrez M, Contreras-Balderas S, Hebert PDN. 2009. Probing diversity in freshwater fishes from Mexico and Guatemala with DNA barcodes. J Fish Biol. 74:377–402.
- Vari RP, Malabarba LR. 1998. Neotropical ichthyology: An overview. In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS. editors. Phylogeny and classification of Neotropical fishes. Porto Alegre: Edipucrs1–11.
- Victor BC. 2007. Coryphopterus kuna, a new goby (Perciformes: Gobiidae: Gobinae) from the western Caribbean, with the identification of the late larval stage and an estimate of the pelagic larval duration. Zootaxa. 1526:51–61.
- Ward RD. 2007. DNA barcoding discriminates spurdogs of the genus Squalus. In: Last PR, White WT, Pogonoski JJ. editors. Descriptions of new dogfishies of the genus Squalus (Squaloidea: Squalidae). Hobart: CSIRO Marine and Atmospheric Research130.
- Ward RD. 2009. DNA barcode divergence within species and genera of birds and fishes. Mol Ecol Resour. 9:1077–1085.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia's fish species. Philos Trans R Soc B Biol Sci. 360:1847–1857.
- Ward RD, Holmes BH, Yearsley GK. 2008. DNA barcoding reveals a likely second species of Asin sea bass (barramundi) (Lates calcarifer). J Fish Biol. 72:458–463.
- Ward RD, Hanner R, Hebert PDN. 2009. The campaign to DNA barcode all fishes. J Fish Biol. 74:329–356.
- Witt JDS, Threloff DL, Hebert PDN. 2006. DNA barcoding reveals extraordinary cryptic diversity in an amphipode genus: Implications for desert spring conservation. Mol Ecol. 15:3073–3082.
- Yassin A, Capy P, Madi-Ravazzi L, Ogereau D, David JR. 2008. DNA barcode discovers two cryptic species and two geographical radiations in the invasive drosophilid Zaprionus indianus. Mol Ecol Resour. 8:491–501.
- Yearsley GK, Last PR. 2006. Urolophus kapalensis sp nov., a new stingaree (Myliobatiformes: Urolophidae) off eastern Australia. Zootaxa. 1176:41–52.