Abstract
Background and aims: DNA barcoding strongly suggests that specimens of the slender codling (Halargyreus johnsonii) from New Zealand and Tasmania belong to a different species to H. johnsonii reported from other areas.
Results: Sequence divergence between the two groups averaged 3.95%, much higher than within-group divergences of 0.03 and 0.02% for specimens, respectively, from New Zealand–Tasmania and from the North Pacific, Atlantic Ocean, and Southern Ocean.
Conclusion: Meristic data for specimens from New Zealand and from the Southern Ocean north of the Ross Sea support the conclusion of two species. DNA barcodes for two sister taxa, Antimora rostrata and Antimora microlepis, show low intra-species (0.3–0.06%) and inter-species (0.23%) divergence.
Introduction
The morid cods (Moridae) are a family of marine benthic cod-like fishes widely distributed in the world's oceans from the Arctic to the Antarctic. The number of morid species (∼100) and genera (17–18) is tentative and there are unsolved taxonomic problems in this group of fishes (Cohen et al. Citation1990). Most species are relatively small and taken as by-catch in trawl fisheries. The slender codling Halargyreus johnsonii Günther Citation1862, also called slender cod or Johnson's cod, is the sole species in the morid genus Halargyreus (Günther Citation1862; Cohen et al. Citation1990; CitationEschmeyer and Fricke 2009). It is a benthopelagic species found on the continental slope at about 450–3000 m with an anti-tropical distribution in the Atlantic and Pacific Oceans (Cohen et al. Citation1990; Gordon et al. Citation1996; Hoff Citation2002; CitationFroese and Pauly 2006). The slender codling reaches a maximum size of c.560 mm standard length (Paulin Citation1983), and while it can be locally abundant it is not targeted but taken as by-catch in deepwater trawl fisheries (Allain et al. Citation2003; McClatchie and Coombs Citation2004). Two other small morid cods, the only members of the genus Antimora (Günther 1878), the longfin cod Antimora microlepis Bean 1890 and the violet cod Antimora rostrata (Günther 1878), are also taken as by-catch in deepwater trawl fisheries; the former in the North Pacific Ocean and the later in all oceans except the North Pacific (Cohen et al. Citation1990). Antimora rostrata is often caught with H. johnsonii, at least around New Zealand (Paulin Citation1983; Paulin et al. Citation2001).
A global campaign to produce DNA barcodes for all marine fishes has been established (Ward et al. Citation2009) and is based on sequence diversity in a single DNA region of the mitochondrial cytochrome c oxidase I gene (COI; Hebert et al. Citation2003). A DNA barcoding survey of New Zealand and Antarctic fishes has examined c.500 species to date. Most species for which multiple specimens have been sequenced for COI are characterized by low intra-specific sequence divergence ( < 0.5%) and higher intra-generic divergence (>4%) (P.J. Smith and D. Steinke, 2010), and are typical of intra-specific and inter-specific COI sequence divergences reported in marine fishes (Ward et al. Citation2005; Steinke et al. Citation2009b). Here, we report high sequence divergence values observed among specimens of H. johnsonii from around New Zealand and the Southern Ocean indicating the presence of two species. On the other hand, the two sister taxa A. microlepis and A. rostrata showed very little divergence both within and between species.
Materials and methods
Samples
Specimens of Halargyreus and Antimora caught on commercial and research vessels around New Zealand and Tasmania, and in the Southern Ocean, were frozen whole at sea for onshore processing—except those on an IPY-CAML Tangaroa survey, which were tissue sampled at sea. Muscle samples from fresh or thawed specimens were stored in 90% ethanol and many of the specimens were then fixed in 10% formalin, prior to storage in 50% isopropanol or 70% ethanol and registration into one of three collections: the Museum of New Zealand Te Papa Tongarewa; the Australian National Fish Collection at Hobart, Australia; and the Australian Antarctic Division, Hobart, Australia. Collection locations and specimen reference numbers are listed in Appendix 1 and shown in .
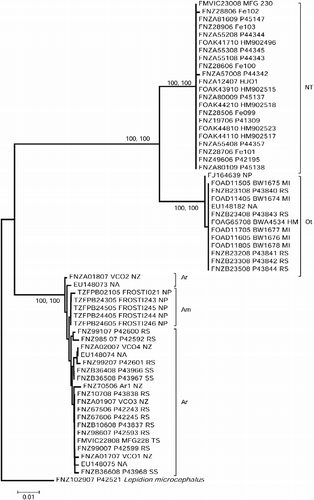
Figure 1. Kimura 2-parameter (K2P) distance neighbor-joining tree of 35 COI sequences from H. johnsonii (NT = New Zealand–Tasmania; Ot = other regions), five sequences from A. microlepis (Am) and 21 sequences from A. rostrata (Ar). BOLD accession numbers are given for each specimen followed by a sample ID, and location abbreviation (HM, Heard and McDonald Islands; MI, Macquarie Island; NA, North Atlantic Ocean; NP, North Pacific Ocean; NZ, New Zealand; RS, Ross Sea; SS, South Sandwich Islands); five sequences are taken from Genbank, H. johnsonii: FJ164639 and EU148182, A. rostrata: EU148703/74/75. The tree has been rooted with the morid outgroup L. microcephalus. Numbers at nodes are bootstrap percentages (>70%) after 1000 replicates, based on distance and Bayesian posterior probability values (>90%); scale bar is for K2P distance.
DNA extraction, amplification, sequencing, and data analysis
DNA was extracted from sub-samples of muscle tissue taken from reference specimens of H. johnsonii and A. rostrata using an automated glass fiber protocol (Ivanova et al. Citation2006). The 650 bp barcode region of COI was amplified under standard conditions (Ivanova et al. Citation2007). PCR products were visualized on a 1.2% agarose gel E-GelH (Invitrogen, Carlsbad, California, US) and sequenced in both directions, using the sequencing primers M13F and M13R (Ivanova et al. Citation2007), and the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Inc., [ABI], Carlsbad, California, US) on an ABI 3730 DNA analyzer following the manufacturer's instructions. Specimen and collection data, sequences, and trace files are available on the Barcode of Life database (BOLD; http://www.boldsystems.org; Ratnasingham and Hebert Citation2007) in the project file “Halargyreus”; BOLD accession numbers are given in and Appendix 1. Two additional COI sequences of H. johnsonii specimens from the North Pacific Ocean and North Atlantic Ocean were taken from Genbank (accession numbers: FJ164639 and EU148182, respectively). COI sequences from A. microlepis from the Canadian North Pacific Ocean and A. rostrata from geographic regions other than New Zealand, the Ross Sea, and the South Sandwich Islands were obtained from BOLD and Genbank; specimen reference numbers are given in and Appendix 1. A sequence divergence value set at 10 × the average within species divergence has been recognized as effective for detecting cryptic species in birds (Tamura et al. Citation2007).
Initial neighbor-joining clustering used the BOLD Management & Analysis System. Phylogenetic relationships were explored with distance methods using PAUP* v4b10 (Swofford Citation2003), with support for each internode evaluated by 1000 bootstrap pseudo-replicates (Felsenstein Citation1985). Bayesian phylogenetic analyses were estimated with MrBayes v3.1.2 (Ronquist and Huelsenbeck Citation2003). The TrN substitution model with equal base frequencies (Tamura and Nei Citation1993) was selected as the best-fit nucleotide substitution model by jModeltest v0.1.1 (Posada Citation2008) using the Bayesian information criterion. Four simultaneous Monte Carlo Markov chains were run for one million generations, saving the current tree every 1000 generations. A 50% majority-rule consensus tree was created with a burn-in value of 1000 (i.e. the first 1000 trees were discarded). Another morid, Lepidion microcephalus Cowper 1956, in the genus Lepidion Swainson 1838 was used to root the trees.
Meristic characters
Fifteen meristic characters () were counted in six of the reference Halargyreus specimens from New Zealand and six of the reference specimens from the Southern Ocean. These specimens had yielded COI sequences and are registered and held in the National Fish Collection at the Museum of New Zealand Te Papa Tongarewa. Counts followed standard methods (Hubbs and Lagler Citation1949) and were compared with results from Paulin (Citation1983) and Gon and Heemstra (Citation1990).
Table I. Nucleotide distance (% Kimura two-parameter) within and between species of H. johnsonii, A. rostrata, A. microlepis, and L. microcephalus.
Results
The Bayesian and neighbor-joining tree revealed two well-supported (100%) clades among the H. johnsonii specimens: New Zealand–Tasmania and all other regions. Sequences from the North Pacific Ocean and North Atlantic Ocean specimens were very similar to or identical with those from the Southern Ocean (). Sequence divergence values were high in the pooled H. johnsonii dataset (3.95%), but low within New Zealand–Tasmania (0.03%), within the Southern Ocean (0%), and within all regions excluding New Zealand and Tasmania (0.02%) (). Sequence divergences were low both within (0.32 and 0.06, respectively) and, surprisingly, between (0.23%) the two sister taxa, A. rostrata and A. microlepis ().
Of the 15 meristic characters examined in specimens of Halargyreus from New Zealand and from north of the Ross Sea, two characters—the number of rays in the second dorsal fin (D2) and the anal fin (A)—distinguished New Zealand specimens (D2, 56–59; A, 47–51) from Southern Ocean specimens (D2, 50–54; A, 42–46) ().
Table II. Meristic counts from H. johnsonii from the Southern Ocean just north of the Ross Sea and New Zealand, along with values given in Gon and Heemstra (Citation1990) and Paulin (Citation1983).
Discussion
For a sequence divergence value set at 10 × , the average intraspecific variation has been recognized as effective for detecting cryptic species in birds (Hebert et al. Citation2004). A comparison of 35 commercially harvested species of Indo-Pacific fishes found in Australian and South African waters used a sequence divergence of about 10 × , the average within-species value as a screening threshold to detect likely overlooked species, and found that on this basis current taxonomic knowledge substantially underestimated species diversity in marine fishes in this region (Zemlak et al. Citation2009).
The degree of differentiation between specimens of H. johnsonii from New Zealand–Tasmania and elsewhere is about 20-fold that within either of these two regions. This high sequence divergence is typical of congeneric species (Steinke et al. Citation2009a; Ward et al. Citation2009; Zemlak et al. Citation2009) and indicates the presence of a cryptic species within what is currently recognized as the single species H. johnsonii. An earlier DNA barcoding study comparing North Atlantic and Australasian fishes found no divergence among five specimens of H. johnsonii from the Southern Ocean and one from the North Atlantic Ocean (Ward et al. Citation2008); these sequences are included in the current study.
The COI results coupled with the meristic data strongly suggest that New Zealand specimens of H. johnsonii represent an undescribed species. Specimens from New Zealand can be distinguished from those from the Ross Sea by the number of rays in the second dorsal fin (D2) and anal fin (A). Paulin (Citation1983) reported 48–60 D2 fin rays in 12 specimens from New Zealand, a range greater than found in this study of 12 specimens from New Zealand and the Southern Ocean, but unfortunately did not provide counts for the anal fins. However, (CitationPaulin 1983, p.109) noted that “the New Zealand population of H. johnsonii had on average a greater number of D2 fin rays, more total anal rays, more vertebrae, and more ventral rays than northern hemisphere populations”. Morphometric measurements overlapped considerably and were within the range of species variability, leading to the conclusion that there was “little justification for recognising subspecies” (CitationPaulin 1983, p. 109). Cohen had previously interpreted the variation among northern and southern hemisphere specimens as local intra-specific variation (Cohen Citation1973), but did not comment on this variation in the later catalog of gadiform fishes (Cohen et al. Citation1990). Gon and Heemstra (Citation1990) noted a relatively broad range of meristic counts in H. johnsonii, and remarked that this was apparently due to pooling of several geographical samples from the North and South Atlantic and the Southwest and Southeast Pacific Oceans. Furthermore, they reported differences in D2, A, and V between H. johnsonii from the southern hemisphere (locations not provided) and the North Atlantic Ocean (Gon and Heemstra Citation1990), which parallel the ranges reported here for New Zealand and the Ross Sea specimens ().
The holotype locality for H. johnsonii is Madeira, with the specimen being taken from the stomach of a whiptail gulper eel, Saccopharynx sp. (Günther Citation1862). Given that the one COI sequence from an Atlantic specimen aligned with those from Southern Ocean and North Pacific Ocean specimens (), by taxonomic priority, the New Zealand–Tasmania specimens represent an undescribed species, and are listed here and in BOLD as Halargyreus n.sp. The other two available names, Halargyreus affinis and Halargyreus brevipes, were described from Faroe Bank and off Morocco, respectively, and fall into synonymy with H. johnsoni.
In contrast, the two morid Antimora species revealed very shallow intra-specific and inter-specific divergences (0.1–0.22%) of an extent that typically indicate genetic differentiation within a single species (Ward et al. Citation2005). Small (Citation1981) recognized A. rostrata and A. microlepis as valid species on the basis of a single character, the number and relative size of gill filaments, with 76–90 relatively short gill filaments in A. microlepis from the North Pacific Ocean and 90–103 relatively long gill filaments in A. rostrata from the North Atlantic, Southeast Pacific, and Southern Oceans. Eleven other morphometric characters showed overlap between specimens from the Atlantic and Pacific Oceans (Small Citation1981). Earlier, Iwamoto had considered that there was a single cosmopolitan species of A. rostrata and that A. microlepis, described from just two specimens by Bean (1890), as for five other regional species of Antimora, should be synonymized with A. rostrata (Iwamoto Citation1975). The COI data support the synonymization of A. rostrata and A. microlepis, which, if confirmed, would have important implications for these species that have recently suffered massive declines in abundance due to overfishing (Kulka et al. Citation2003; Devine et al. Citation2006).
DNA barcoding results for another morid cod Pseudophycis bachus show low sequence divergence among specimens within Australia (0.2%) and New Zealand (0%), but high sequence divergence in the pooled data (8.6%), again indicating probable cryptic species (Smith et al. Citation2008).
The apparent discrepancies between morphological descriptions and COI for several species of Moridae indicate that this family requires a taxonomic reappraisal. The number and distributions of species in this family need to be determined, and this would be assisted by a detailed molecular phylogeny that included additional specimens from the North Atlantic and Pacific Oceans.
Acknowledgements
The authors are grateful to two anonymous referees for constructive comments on the manuscript. New Zealand Halargyreus and Antimora specimens were collected by one of the authors (P.M.) and Ross Sea specimens by two of the authors (P.M. and A.S.) aboard the NIWA Research vessel Tangaroa. Specimens from around Heard and McDonald Islands and from Macquarie Island were collected by Australian Fisheries Management Authority observers.
Declaration of interest: New Zealand authors were supported by research funded by the New Zealand Government under the New Zealand International Polar Year-Census of Antarctic Marine Life Project, and by the Ministry of Fisheries (project GDB200801), and gratefully acknowledge the Ministry of Fisheries Science Team and Ocean Survey 20/20 CAML Advisory Group (Land Information New Zealand, Ministry of Fisheries, Antarctica New Zealand, Ministry of Foreign Affairs and Trade, and National Institute of Water and Atmospheric Research Ltd). D.S. was supported by funding from the Alfred P. Sloan Foundation to MarBOL and by Genome Canada, through the Ontario Genomics Institute.
References
- Allain V, Biseau A, Kergoat B. 2003. Preliminary estimates of French deepwater fishery discards in the Northeast Atlantic Ocean. Fish Res. 60:185–192.
- Cohen DH. 1973. The gadoid fish genus Halargyreus (Family Eretmophoridae) in the southern hemisphere. J R Soc NZ. 3:629–634.
- Cohen DM, Inada T, Iwamoto T, Scialabba N. 1990. Gadiform fishes of the world (Order Gadiformes). An annotatted and illustrated catalogue of cods, hakes, grenadiers and other gadiform fishes known to date. FAO Fish Synop. 125:442 FAO Rome.
- Devine JA, Baker KD, Haedrich RL. 2006. Deep-sea fisheries qualify as endangered. Nature. 439:29.
- Eschmeyer WN, Fricke R. 2009. Catalog of fishes electronic version, 9 September [Online]. Available at: http://research.calacademy.org/ichthyology/catalog/fishcatmain.asp[accessed 10 May 2010].
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 39:783–791.
- Froese R, Pauly D. 2006. FishBase [Online]. Available at: http://www.fishbase.org [accessed 10 May 2010].
- Gon O, Heemstra P. 1990. Fishes of the Southern Ocean. Grahamstown: J.L.B. Smith Institute of Ichthyology462.
- Gordon J, Merrett N, Bergstad O, Swan S. 1996. A comparison of the deep-water demersal fish assemblages of the Rockall Trough and Porcupine Seabight, eastern North Atlantic: Continental slope to rise. J Fish Biol. 49:217–238.
- Günther A. 1862. Catalogue of the fishes in the British Museum. Catalogue of the Acanthopterygii, Pharyngognathi and Anacanthini in the collection of the British Muesum. Cat Fish Br Mus. 4:1–534.
- Hebert PDN, Ratnasingham S, deWaard JR. 2003. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci. 270:S96–S99.
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. 2004. Identification of birds through DNA barcodes. PLoS Biol. 2:e312.
- Hoff GR. 2002. New records of the slender codling Halargyreus johnsonii Günther, 1862 from the eastern Bering Sea. Alsk Fish Res Bull. 9:65–67.
- Hubbs C, Lagler K. 1949. Fishes of the Great Lakes Region. Bloomfield Hills, MI: Cranbrook Press186.
- Ivanova NV, deWaard JR, Hebert PDN. 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes. 6:998–1002.
- Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. 2007. Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes. 7:544–548.
- Iwamoto T. 1975. The abyssal fish Antimora rostrata. Comp Biochem Physiol. 52B:7–11.
- Kulka D, Simpson M, Inkpen T. 2003. Distribution and biology of Blue Hake (Antimora rostrata Guenther 1878) in the Northwest Atlantic with comparison to adjacent areas. J Northwest Atl Fish Sci. 31:299–318.
- McClatchie S, Coombs RF. 2004. Spatial variability of orange roughy around the Northwest Hills on the Chatham Rise, New Zealand. Deep Sea Res. 52:589–603.
- Paulin C. 1983. A revision of the family Moridae (Pisces: Anacanthini) within the New Zealand region. Rec Natl Mus NZ. 2:81–126.
- Paulin C, Stewart A, Roberts C, McMillan P. 2001. New Zealand fishes: A complete guide. Wellington: Te Papa Press, 19, 279.
- Posada D. 2008. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Ratnasingham S, Hebert PDN. 2007. The barcode of life database. Mol Ecol Notes. 7:355–364.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Small GJ. 1981. A review of the bathyal fish genus Antimora (Moridae: Gadiformes). Proc Calif Acad Sci. 42:341–348.
- Smith PJ, Steinke D, McMillan P, McVeagh SM, Struthers CD. 2008. DNA database for commercial marine fish. NZ Aquat Environ Biodivers Rep. 22:62.
- Steinke D, Zemlak TS, Boutillier JA, Hebert PDN. DNA barcoding of Pacific Canada's fishes. Mar Biol. 2009a; 156:2641–2647.
- Steinke D, Zemlak TS, Hebert PDN. Barcoding nemo: DNA-based identifications for the ornamental fish trade. PLoS One. 2009b; 4:e6300.
- Swofford DL. 2003. PAUP*: Phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates Version 4.
- Tamura K, Nei M. 1993. Estimating the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599.
- Ward RD, Zemlak TS, Ines BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia's fish species. Philos Trans R Soc Lond Ser B. 360:1847–1857.
- Ward RD, Costa FO, Holmes BH, Steinke D. 2008. DNA barcoding shared fish species from the North Atlantic and Australasia: Minimal divergence for most taxa but a likely two species for both Zeus faber (John dory) and Lepidopus caudatus (silver scabbardfish). Aquat Biol. 3:71–78.
- Ward RD, Hanner R, Hebert PDN. 2009. The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol. 74:329–356.
- Zemlak TS, Ward RD, Connell AD, Holmes BH, Hebert PDN. 2009. DNA barcoding reveals overlooked marine fishes. Mol Ecol Resour. 9:237–242.
