Abstract
Materials and methods: DNA sequences obtained for the Barcode of Life library in the All Lepidoptera Campaign project Nymphalidae of Central Mexico were analyzed as a test of species limits and to explore possible phylogenetic groupings in the Preponini tribe. Using specimens in the National Insect Collection of the Instituto de Biología of the Universidad Nacional Autónoma de México, 78 specimens were assayed for cytochrome oxidase c subunit 1.
Results: Disregarding the missing data, there were 458 conserved sites, 200 variable sites and 187 parsimony-informative sites. The neighbor-joining and maximum likelihood analyses indicate that none of the three genera of Preponini as currently circumscribed are reciprocally monophyletic. As per species limits, high levels of barcode variation in the Prepona deiphile complex suggest the existence of at least two new endemic species to Mexico. The divergent taxa were escalantiana from the Tuxtlas region in Veracruz, and ibarra from Sierra Madre del Sur in the Pacific states of southern Mexico. The genetic distance in the CO1 fragment between them and the other deiphile populations ranged from 2.7 to 8.0%.
Conclusion: We recommend that morphological data need to be re-examined and that additional molecular data for species ought to be gathered before a particular biogeographic model can be proposed for the group in Mesoamerica.
Introduction
The International Barcode of Life project is an initiative of great appeal to wildlife and resource managers. One of the more active campaigns in this project is the Lepidoptera Barcode of Life (http://www.lepbarcoding.org) because there are considerable number of specimens in collections that could be barcoded, and because adequate protocols are now in place. Additionally, butterflies are quite amenable and prized by people as collectable items, and government agencies should have an efficient method to verify species identification and countries of origin. These goals are achievable through an international initiative in a relatively short time. Because of these considerations, we undertook DNA barcoding of the most attractive component of the lepidopteran fauna of central Mexico, the Nymphalidae. Here, we present the very first results and their interpretation for the tribe Preponini (subfamily Charaxini).
Preponini are large, canopy-dwelling, fruit-feeding nymphalids of the neotropics, most of them with cryptic wing patterns on their underside, and striking iridescent patterns on their dorsal surface. There is as yet very few molecular data at the species level for this group. The Lepidoptera of Mexico have been studied at great geographical detail (i.e. Luis-Martínez et al. Citation2003), but a revision using a new suite of characters such as mitochondrial DNA sequences has not yet been produced. In our paper, we present a preliminary phylogenetic analysis of closely related species in the subfamily Charaxinae, tribe Preponini Rydon 1971, mostly from the west of the Isthmus of Tehuantepec, using cytochrome oxidase c subunit 1 (cox1) sequences.
The tribe Preponini is currently composed of 21 species in four genera, distributed mainly in the Neotropical Region. In Mexico, there are representatives of 11 species in three genera. The current taxonomy (CitationSavela 2010) of this tribe acknowledges these: Prepona (seven species, of which four are in Mexico), Archaeoprepona (eight species, of which five are in Mexico), Agrias (five species, of which two are in Mexico) and Noreppa (one species, none in Mexico). Anaeomorpha, from South America, is sometimes recognized as a monotypic genus in this tribe, too.
As updated by CitationWahlberg and Brower (2009), the sister group of Preponini are members of the Anaeini tribe. This is also in accordance with a recent cladistic analysis of morphological characters for the subfamily Charaxinae (CitationMarconato 2008), in which, except for Anaeomorpha splendida, Preponini is monophyletic. At the generic level, Archaeoprepona and Prepona were polyphyletic, since Archaeoprepona should include the monotypic genus Noreppa, and Prepona should include Agrias, to be monophyletic. With these findings, we chose a member of Anaeia as the sister group for our analysis, and as additional outgroups we included seven other Nymphalids from the Barcode of Life Data Systems (BOLD) public database.
Many species of Preponini have a number of described subspecies. One such example is the Prepona deiphile complex reviewed by Llorente-Bousquets et al. (Citation1992), in which differentiating isolated populations occurring in various mountainous ranges are given subspecies status according to a particular morphological interpretation and a previously chosen biogeographical hypothesis, which is understood as a “major vicariant pattern in southern Mexico” with one component in the east and southeast part of the country, and the second component in the south and west (Llorente-Bousquets et al. Citation1993; Vargas-Fernández et al. Citation2006). DNA barcodes can help by providing data to test this biogeographic hypothesis.
Materials and methods
Samples
Tissue samples were collected from specimens deposited in the National Insect Collection at the Instituto de Biología, Universidad Nacional Autónoma de México (CNIN-LEP, IBUNAM). Legs were removed and sent for genetic analysis to the Biodiversity Institute of Ontario at the University of Guelph (Guelph, Ontario, Canada). The specimens were photographed and entered into the database of the Unidad de Informática de la Biodiversidad (Instituto de Biología, UNAM; http://www.unibio.ibiologia.unam.mx), and subsequently into BOLD (http://www.boldsystems.org). Most of the subspecies described for this tribe in Mexico were represented in this data set.
DNA extraction, PCR amplification and sequencing
The samples were subjected to the regular laboratory procedures used in the All-Lepidoptera Campaign for lysis and DNA extraction, but a “minibarcode” amplification was undertaken to account for the specimens' age (collected between the 1960s and 1990s). Rather than a PCR to amplify a 650 bp fragment in a single reaction for each sample, two smaller fragments were amplified in separate PCR reactions. Only 10 samples failed to amplify for both fragments, while only one of the two fragments was successfully amplified for about 24 of the 84 specimens. Here, we only analyze the specimens belonging to the Tribe Preponini (Archaeoprepona, Prepona, and Agrias spp.) with a minimum sequence length of 307 bp (68 samples, of which 36 were 658 bp in length, 22 were 407–602 bp and 10 were 307 bp). All species/populations were represented by at least two full barcodes. The sequences obtained were deposited in BOLD and GenBank ().
Table I. Specimen data used in the present study.
Alignment and phylogenetic reconstruction
The 68 DNA sequences were aligned in ClustalX (Thompson et al. Citation1997) and then we used the neighbor-joining (NJ) tree tool with the Kimura two-parameter substitution model (Kimura Citation1980) in BOLD with node support estimated through 100 bootstrap replicates. We translated our nucleotide sequences into amino acids in DnaSP 5.10.01 (Librado and Rozas Citation2009) and no stop codons were encountered. For outgroups, we chose a sequence from Costa Rica of Anaea aidea, already available in BOLD and seven other outgroups (see , for full data and GenBank accession numbers), following Peña and Wahlberg (Citation2008).
Disregarding missing data, there were 458 conserved sites, 200 variable sites and 187 parsimony-informative sites. Nucleotide composition was 39.2% T, 16.5% C, 30% A and 14.3% G; that is, within the expected range for a coding gene (Saccone et al. Citation1999; Junqueira et al. Citation2004).
We selected the best-fit substitution model for our alignment using jModelTest 0.1.1 (Posada Citation2008), and then used those parameters for a maximum likelihood (ML) phylogenetic analysis in PAUP* 4b10 (Swofford Citation2003) using the NJ tree as the start tree for a heuristic search. The settings from the best-fit model (TIM2+I + G) selected by the Bayesian information criterion used in PAUP* were: Lset base = (0.3001 0.1425 0.1382 0.4192) nst = 6 rmat = (10.2850 24.5568 10.2850 1.0000 84.4402 1.0000) rates = gamma shape = 1.1510 ncat = 4 pinvar = 0.5880. Four trees were obtained with the same likelihood score and were summarized through a majority-rule consensus tree. For visualization, rooting the tree and labeling the tips, we used FigTree (CitationRambaut 2007). For constructing the map in using a niche model, we used the program DIVA-GIS (CitationHijmans et al. 2005).
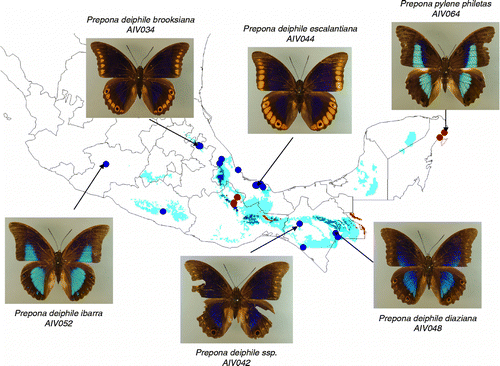
Figure 1. Prepona deiphile species group. Distributional ranges of the P. deiphile forms in Mexico as predicted with the DIVA-GIS program. Some representative specimens used in this study are depicted. Blue dots and shades represent localities and ranges for deiphile forms, while red dots and shades for pylene.
Results and discussion
Genetic distance analysis
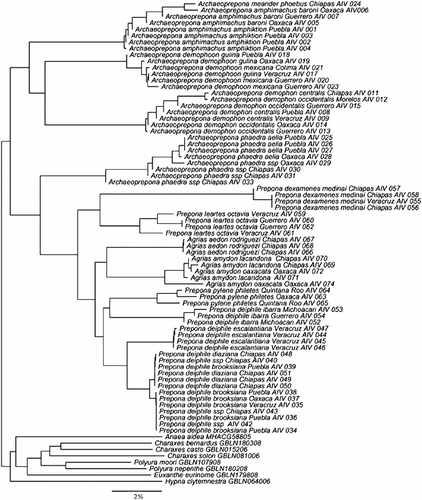
For all the ingroup taxa, within-species distances ranged from 0 to 8.004. Within species level designations, distances above 2.87 and up to 8.0 were observed for Archaeoprepona phaedra (five out of 15 comparisons), P. deiphile (84 out of 171 comparisons) and Prepona pylene philetas (one out of one comparison). There were three peaks in the pairwise comparisons within species: one at 0.25, another at 3.12 and a third at 5.25. The NJ distance tree is shown in .
Preliminary phylogenetic reconstruction
No previous genetic distance trees have been produced for Preponini at this level. The NJ tree indicated that Archaeoprepona is perhaps a polyphyletic genus, with A. phaedra more closely allied with the Prepona–Agrias group () than with the other three Archaeoprepona species. However, the ML tree (not shown) did not recover Archaeoprepona as monophyletic at any level. Prepona was found to be paraphyletic because it included Agrias in both the NJ and the ML trees, a result supported by a previous cladistic analysis (CitationMarconato 2008). The Prepona–Agrias group did not show the same topology in the NJ and the ML analyses, but the same lower groups were recovered by both analyses. A comprehensive phylogenetic reconstruction of the Preponini will require greater taxon sampling and more informative characters.
Species limits
On the other hand, our results were consistent for distance and phylogenetic criteria regarding species-level groupings. The same groups can be recognized using either the >2% genetic distance criteria for species or the reciprocal monophyletic criterion in the likelihood analysis. Except for Archaeoprepona meander, all the other four Archaeoprepona species were recognized by both criteria, but A. phaedra deserves a more detailed geographical and phylogenetic analysis to better understand its taxonomic status.
Additional interesting results are emerging in the Prepona group. Prepona laertes and Prepona dexamenes were sister taxa. The deiphile group was polyphyletic; first, deiphile ibarra of the Sierra Madre del Sur was the sister group of P. pylene, although the two were separated by a considerable genetic distance. This relationship was suggested originally by Beutelspacher (Citation1982). Another segregate outside the main deiphile subspecies was deiphile escalantiana, which occurs only in Los Tuxtlas, Veracruz in the Gulf of Mexico coast, separated by a genetic distance of more that 2.97% from the main deiphile forms. The remaining populations from the cloud forests of the Sierra Madre Oriental and Chiapas form two closely related, perhaps incipiently differentiating forms.
Taxonomic recommendations
The cox1 data collected thus far suggest that the current taxonomy of Preponini needs revision, and a closer scrutiny of morphological, genetic, and behavioral data will produce a better understanding and classification of Preponini. We would like to point out specific suggestions for lepidopterists to consider:
| • | (a) The possible merging of A. meander and Archaeoprepona amphimachus. The genetic distance between specimens from both species was negligible, and our observations of the available specimens indicate that there are very subtle and subjective morphological differences between these two species. | ||||
| • | (b) The merging of Agrias into Prepona. Because this was also suggested by the cladistic analysis of CitationMarconato (2008), and recognized in the literature by previous authors, the mitochondrial DNA and new morphological interpretations could support this change. | ||||
| • | (c) The upgrade of ibarra (Beutelspacher Citation1982) to species status. By giving it only a subspecies status, some differentiation was recognized but this form is widely geographically separated, together with lambertoana from the rest of the deiphile forms, which seem to match perfectly with the mitochondrial separation. There also seem to have less sexual dimorphism than that in other forms of deiphile. | ||||
| • | (d) The elevation of escalantiana (Stoffel and Mast 1973) to the species level. This form is strikingly similar to Prepona xenagoras of Peru, especially in the presence of ocelli with orange rectangles on the external part of veins of the dorsal part of their wings. This character then shows a leapfrog geographic pattern in which perhaps escalantiana and xenagoras have retained a primitive character. Species escalantiana also has another important morphological characteristic that differs from the rest of the deiphile forms: very little sexual dimorphism in coloration exhibited by most forms of deiphile. | ||||
Biogeographic patterns
Although a phylogeographical analysis would be premature with the present data for the species groups represented in this study, it is clear that the vicariant model of eastern and western forms north of the Isthmus of Tehuantepec proposed to explain differentiation in these series of species populations does not hold as a general pattern. None of the four Archaeoprepona species included in this study presented a clear significant differentiation between eastern and western forms that would give validity to the subspecies proposed as evidence of this pattern. The same could be said among the Prepona species represented in this study, and particularly in the P. deiphile group in which potentially new endemic species revealed by the cox1 data could deserve recognition. These results suggest that the biogeographic history of Preponini in this region is more complicated and involves perhaps a series of dispersion and vicariant events at different ages and opportunities, with more than a single model applying for the species.
GenBank accession numbers for outgroups: Anaeia aidea MHACG58805, GU333743; Charaxes bernardus GBLN180308, EF534101; Charaxes castor GBLN015206, AY090219; Charaxes solon GBLN081006, DQ810197; Polyura moori GBLN107908, EU528325; Polyura nepenthe GBLN180208, EF534102; Euxanthe eurinome GBLN179808, EU141357; Hypna clytemnestra GBLN064006, DQ338574.
Acknowledgements
The authors thank Evgeny Zakharov, Marianne N. A. Iskandar, and Suresh Naik for laboratory work at the Biodiversity Institute of Ontario, University of Guelph. They also thank Matthew J. Miller, Postdoctoral Fellow at the Smithsonian Tropical Research Institute in Panama, for assistance with the analysis. Valuable advice was also received from Sergios-Orestis Kolokotronis and David Gernandt.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Beutelspacher C. 1982. Una nueva subespecie del género Prepona Boisduval (Lepidoptera, Nymphalidae) de México. An Inst Biol Univ Nac Autón Méx Ser Zool. 46 1: 367–370.
- Hijmans RJ, Guarino L, Jarvis A, O'Brien R, Mathur P, Bussink C, Cruz M, Barrantes I, Rojas E. 2005. DIVA-GIS version 5.2. Available at http://www.diva-gis.org.
- Junqueira AC, Lessingera AC, Torres TT, Rodrigues da Silva F, Vettorec AL, Arrudad P, Azeredo-Espina AML. 2004. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae). Gene. 339:7–15.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.
- Llorente-Bousquets J, Luis-Martinez A, González-Cota L. 1992. Diferenciación de Prepona deiphile en Mesoamérica y descripción de dos subespecies nuevas (Lepidoptera: Nymphalidae). Trop Lepid. 3:109–114.
- Llorente-Bousquets J, Descimon H, Johnson K. 1993. Taxonomy and biogeography of Archaeoprepona demophoon in Mexico with description of a new subspecies (Lepidoptera: Nymphalidae: Charaxinae). Trop Lepid. 4:31–36.
- Luis-Martínez A, Llorente J, Vargas IF, Warren AD. 2003. Biodiversity and biogeography of Mexican butterflies (Lepidoptera: Papilionoidea and Hesperioidea). Proc Entomol Soc Wash. 105:209–224.
- Marconato G. 2008. Análise cladística de Charaxinae Guenée (Lepidoptera, Nymphalidae). PhD Thesis, Universidade de São Paulo, São Paulo, Brazil, p 180. Available at http://www.teses.usp.br/teses/disponiveis/41/41133/tde-02032009-154826.
- Peña C, Wahlberg N. 2008. Prehistorical climate change increased diversification of a group of butterflies. Biol Lett. 4:274–278.
- Posada D. 2008. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Rambaut A. 2007. FigTree, A Graphical Viewer of Phylogenetic Trees. Available at http://tree.bio.ed.ac.uk/software/figtree.
- Saccone C, de Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene. 238:195–209.
- Savela M. 2010. Markku Savela's Lepidoptera and Some Other Life Forms – Charaxinae. Available at http://www.funet.fi/pub/sci/bio/life/insecta/lepidoptera/ditrysia/papilionoidea/nymphalidae/charaxinae/index.html (accessed on 11 March 2010).
- Swofford DL. 2003. PAUP*: Phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876–4882.
- Vargas-Fernández I, Trujano M, Llorente-Bousquets J, Luis-Martínez A. 2006. Patrones de distribución de las subfamilias Ithomiinae, Morphinae y Charaxinae (Lepidoptera: Nymphalidae). In: Morrone JJ, Llorente-Bousquets J. editors. Componentes bióticos principales en la entomofauna mexicana. México, DF: Las Prensas de Ciencias, UNAM. p 867–943.
- Wahlberg N, Brower AVZ. 2009. Tree of Life Web Project: Charaxinae Doherty 1886. Available at http://www.tolweb.org/Charaxinae (accessed on 9 May 2010).