Abstract
Background and aim. The avocado stem weevil Copturus aguacatae is an important pest in avocado plantations. Its presence hinders the production and marketing of avocado in Mexico, the largest avocado producer worldwide. Biological control through pheromone synthesis, a strategy favored over chemical control in crops, is currently limited by difficult field identification of this species.
Materials and methods. Using DNA barcoding, we examine the patterns of genetic variation of C. aguacatae in avocado trees in Mexico to help facilitate its identification and biological control.
Results. We show that there is one single species of avocado stem weevil throughout the sampled sites in Mexico. Overall, haplotype diversity is high, with Oaxaca forming one distinct group and all other sampled populations are admixed irrespective of geographic origin.
Conclusion. The results suggest that high gene flow is maintained in this species and that a global strategy for biocontrol can be designed and implemented throughout the sampled range.
Introduction
Mexico is the world's largest avocado producer, growing more than 30% of the world's total production and providing over 40% of the world's avocado exports (Lamb Citation2006). Avocado is grown in 16 Mexican states, with Michoacán and Puebla, which lie within Mexico's “Avocado Belt,” being the largest producers (Lamb Citation2006). The avocado stem weevil, Copturus aguacatae Kissinger, 1957, is a native pest to Mexico and can be found as far South as Guatemala. It destroys both the avocado tree and sometimes the fruit (Ochoa and Santacruz Citation1999), making this insect pest a major limiting factor in the production and the marketing of avocados both nationally and internationally (Téliz Citation2000).
As of yet, there is no cost-effective, successful control for these weevils. Most chemical methods of control require knowledge of the weevil's life cycle to determine when to apply insecticides, which varies depending on altitude and climate (Talavera and Padilla Citation2003). Moreover, the frequent application of insecticides has environmental consequences, may impede biological control for other pests, and can interfere with harvest activities (Schilmann et al. Citation2009). The only known predators of the insect are birds, which do not reduce its population enough to curtail the damage caused by these insects (Coria et al. Citation2007). An effort is currently underway to isolate a pheromone of the insect, which would attract adults of C. aguacatae to a trap laced with pheromone with minimal detrimental economic and ecological impacts. Using pheromones as a control for harmful insects has successfully been done with its sympatric species Anthonomus eugenii, the pepper weevil (Eller et al. Citation1994) and is a widespread practice for many other pests. However, before being able to implement pheromone control, it must be confirmed that C. aguacatae is indeed one species and not several—in which case a pheromone would be isolated only for the most populous species (CitationCibrián Tovar 2008). Although hypothesized to be a single species in avocado trees in Mexico (CitationCibrián Tovar 2008), field identifications are equivocal for C. aguacatae due to its natural morphological variation, suggesting that there may be more than one species present throughout the country.
Traditional methods of identification based on morphology are successful for identifying insect relationships at higher taxonomic levels, but genetic methods of identification are often necessary to elucidate the relationships among insect species and subspecies (Caterino et al. Citation2000). DNA barcoding is an effective way of distinguishing species when morphological field identification is difficult, as is the case with C. aguacatae. The mitochondrial gene cytochrome c oxidase I (Cox1) is widely used for DNA barcoding given its relatively high mutation rate resulting in patterns unique to animal species, including insects (Hajibabaei et al. Citation2005). Here, we present the first DNA sequence data from the Cox1 gene of C. aguacatae and discuss the implications of barcoding for identifying insect pest species on important crops, such as avocado.
Materials and methods
Samples
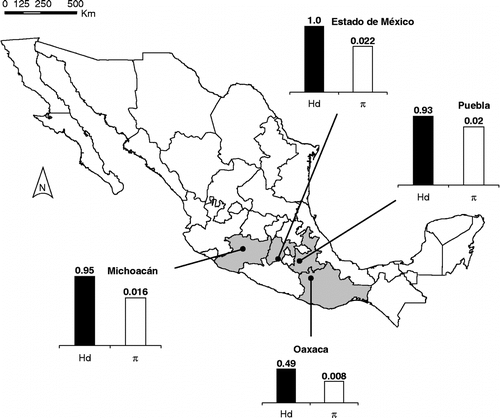
Copturus aguacatae specimens were collected as both larvae and adults from four different localities in different states during May–June 2008: Villa de Chilapa de Diaz, Oaxaca State; Huaquechula, Puebla State; Ziracuaretiro, Michoacán State; and Coatepec de Harinas, Mexico State (hereafter Estado de México) (). Insects were removed from the avocado plants on which they were feeding and placed into vials containing 100% v/v ethanol.
Laboratory procedures
DNA was extracted from ethanol-preserved specimens using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. For adult specimens, the rear right leg was removed and then crushed with a rotor-stator homogenizer to expose as many cells as possible. Larval specimens were cut in half and then crushed with a micropipette tip. An approximately 850 bp DNA fragment was obtained by PCR amplification of Cox1 in a 25 μl reaction volume containing 2.5 μl buffer A (containing MgCl2), 2.5 μl dNTPs, 1.0 μl of each primer, 0.1 μl BioReagents Taq DNA polymerase (Fisher Scientific, Pittsburgh, PA, USA), and 1.0 μl DNA template. We used the primers C1-J-2183 (Jerry) 5′-CAACATTTATTTTGATTTTTTGG-3′ and TL2-N-3014 (Pat) 5′-TAATATGGCAGATTAGTGCATTGGA-3′ (Simon et al. Citation1994). Thermal cycling conditions involved an initial denaturation step at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, an annealing temperature of 53°C for 1 min, and an extension temperature of 70°C for 2 min and 15 s. This was followed by an additional 10 min at 70°C. All PCR reactions were carried out on a Mastercycler ep thermal cycler (Eppendorf, Hauppauge, NY, USA). PCR products were visualized on a 1% w/v agarose gel stained with SYBR Safe DNA gel stain (Invitrogen, Carlsbad, CA, USA). We purified the PCR product using an Agencourt AMPure PCR purification kit (Beckman Coulter Genomics, Brea, CA, USA). Cleaned PCR products were cycle sequenced with BigDye Terminator Cycle Sequencing kit v3.1 and sequenced on a 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
Data analysis
Sequence chromatograms were edited in Sequencher 4.2 (Gene Codes Corp, Ann Arbor, MI, USA). Sequences were trimmed to approximately 780 bp and then aligned in Geneious Pro 4.6.5 (Biomatters, Auckland, New Zealand; http://www.geneious.com) using MUSCLE 3.6 with a maximum number of eight iterations (Edgar Citation2004). The final alignment included sequences from individual insects belonging to C. aguacatae (n = 27). The nucleotide sequences along with voucher specimen information and collection locality geographical coordinates were deposited in the Barcode of Life Data Systems (http://www.boldsystems.org; CitationRatnasingham and Hebert 2007) under project code “WEEMX” (Avocado weevils of Mexico) and, subsequently, in GenBank under accession numbers HQ599817-43.
The phylogenetic relationships among individuals were investigated using maximum likelihood in the Pthreads built of RAxML 7.2.6 (Stamatakis Citation2006; Stamatakis and Ott Citation2008). Fifty maximum likelihood searches were launched using individual maximum parsimony stepwise-addition starting trees and the General Type Reversible (GTR) substitution model (Lanave et al. Citation1984) with among-site rate heterogeneity accounted for using four rate categories of the Γ distribution (Yang Citation1994). Node support was estimated using 1000 nonparametric bootstrap pseudo-replicates (Felsenstein Citation1985).
Synonymous and nonsynonymous mutations between populations were counted by eye in Se-Al 2.0a11 (http://tree.bio.ed.ac.uk/software/seal). We measured the extent of DNA polymorphism in DnaSP 5.10.01 (Librado and Rozas Citation2009; http://www.ub.edu/dnasp), namely nucleotide diversity (π), the number of haplotypes (h), and haplotype diversity (Hd) (Nei Citation1987). Genetic differentiation across the sampled regions of Mexico was assessed with Hudson et al. (Citation1992) HST and KST tests in DnaSP, while among sampling localities, it was quantified by means of FST in Arlequin 3.5.1.2 (Excoffier and Lischer Citation2010; http://cmpg.unibe.ch/software/arlequin35). FST pairwise distances were calculated using the Tamura–Nei correction (Tamura and Nei Citation1993) that distinguishes between transition and transversion rates, and among-site rate heterogeneity modeled by the Γ distribution (α shape parameter set at 0.02 as estimated in RAxML). Statistical significance in genetic differentiation tests was assessed through 50,175 permutations.
Results
Of 40 total C. aguacatae samples collected, only 27 were successfully sequenced. C. aguacatae haplotypes formed four well-supported groups (). All but one Oaxacan individuals formed a strongly supported “Group I” (97% bootstrap support), suggesting that there is considerable divergence separating the Oaxacan population from populations from the other sampled states. Eight identical sequences from Oaxaca are grouped together (90% bootstrap support) within Group I. The remaining three groups, while well supported, are admixed and composed of individuals from all different localities.
Figure 2. Maximum likelihood phylogenetic tree. Values on the internode branches indicate clade support calculated from 1000 bootstrap pseudo-replicates; only values above 90% are shown. Filled circles indicate strongly supported nodes (>97%). The tree is midpoint-rooted. Log likelihood = − 1282.081877.
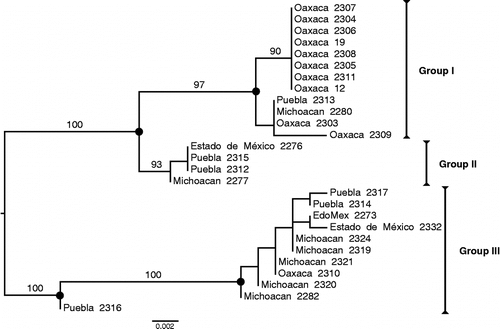
Groups II and III each have one unique nonsynonymous substitution (Ser–Gly and Val–Ile, respectively) separating them from the other groups. Serine is a hydrophilic amino acid which can participate in hydrogen bonding, while glycine, the smallest amino acid, is nonpolar and cannot participate in hydrogen bonding (CitationIUPAC–IUB Joint Commission on Biochemical Nomenclature 1983). Valine is a small hydrophobic amino acid, and isoleucine has branched hydrocarbon side chains; however, these side chains are not reactive (CitationIUPAC–IUB Joint Commission on Biochemical Nomenclature 1983). There are also 30 synonymous substitutions that divide the three larger clades into smaller groups. A full list of intraspecific substitutions is available from the authors upon request.
We uncovered a great extent of genetic variability (π = 0.01858; Hd = 0.897) and population subdivision (H-ST = 0.18402, p < 0.001; KST = 0.23089, p = 0.0088) across C. aguacatae, with 14 haplotypes and 34 variable sites (). The Oaxaca population was markedly distinct by having nine private haplotypes and showing significant differentiation with respect to Michoacán (FST = 0.53965, p = 0.01018) and Estado de México (FST = 0.63563, p = 0.00544), but not when compared with Puebla (FST = 0.26989, p = 0.09245). Oaxaca shared one haplotype with Puebla and two haplotypes with Michoacán. The other three populations were not significantly differentiated in a pairwise fashion (p>0.31). Oaxaca showed the lowest nucleotide (π = 0.00773) and haplotype (Hd = 0.49091) diversity, while Estado de México was the most variable population (π = 0.02248), with all of its three individuals having distinct haplotypes (Hd = 1.0) and two of its three haplotypes being private.
Table I. Summary statistics for Cox1 sequences of avocado weevils.
Discussion
These findings indicate that, despite natural morphological variation, avocado weevils distributed throughout the sampling sites belong to C. aguacatae—confirming observations based on both morphological and life cycle studies by CitationCibrián Tovar (2008). In our phylogenetic analysis, while almost all Oaxacan specimens grouped together, the remaining specimens from Michoacán, Estado de México, and Puebla intermingled, forming Groups I and III. This admixture could be expected with individuals from Estado de México and Puebla, as the two locations are fairly close to one another. However, Michoacán is over 500 km from Estado de México, the nearest locality, yet is still included in Groups I and III. This suggests that none of these populations has any informative unique substitutions separating them from one another, which could have indicated separate evolutionary trajectories. Furthermore, all of the groups (I–III) are genetically close enough to indicate that they are all one species.
Counting substitutions by eye revealed that there are only two amino acid substitutions within C. aguacatae. The first (Ser to Gly), which is unique to Group III, could alter the protein's tertiary structure and potentially its function, too, due to the different sizes and polarities of the two amino acids. However, due to a lack of experimental data and the incomplete nature of our coding sequence, we cannot predict unequivocally the effects of amino acid replacements on protein structure and function. The second substitution (Val to Ile) is specific to Group IV. Although these amino acids have different structures, isoleucine is typically considered interchangeable with valine in proteins (Brosnan and Brosnan Citation2006). This substitution is, therefore, not expected to alter the function of the protein. Furthermore, one amino acid difference is not enough as evidence to suggest that Groups III and IV are a different species from Groups I and II.
Thirty synonymous substitutions were detected among all C. aguacatae individuals. Assuming that valid species—allopatric or not—are most probably separated by adaptive genetic divergence expressed by protein sequence differences, and given the partial gene sequence data presented here as preliminary information, we view synonymous polymorphisms as natural intraspecific variation and the existence of private sites and haplotypes as the result of population separation and isolation. Our mitochondrial DNA sequence results do not support the existence of multiple species of C. aguacatae in the regions examined in Mexico. Ongoing work extending to nuclear loci (e.g. 28S rDNA) shows virtually identical genetic profiles across the sampled populations (data not shown), therefore, supporting our results based on mitochondrial DNA.
Research on avocado stem weevil attractants, as in other weevil species (Calyecac-Cortero et al. Citation2004), will require a growing number of insects of the same sex and age for behavior, extracts, and bioassays. Based on our results, we are now able to employ populations of C. aguacatae from different locations in Mexico for tests and development of attractants. It remains of paramount importance in the field of economic and applied entomology to have a tool such as DNA barcoding for the unambiguous identification of species of interest. In particular, identifying agriculturally important pests can be a challenging task hampered by many factors, including misidentification in the literature, misplacement at higher taxonomic levels, and morphologically cryptic species (Pogue and Simmons Citation2008). All of these issues can be addressed with the use of DNA barcoding and—where needed—nuclear DNA sequencing for species delimitation and the disentangling of population-level processes, such as demographic changes and colonization of novel areas and habitats.
Declaration of interest: This work was funded by the Dirección General de Sanidad Vegetal of Mexico through a grant conferred on J.C.T. The laboratory work was supported by the Cullman Program in Molecular Systematics at the American Museum of Natural History (AMNH) and the New York Botanical Garden (NYBG), the Alfred P. Sloan Foundation to the AMNH DNA Barcoding Initiative for Conservation, and the AMNH Sackler Institute for Comparative Genomics. R.C.E. was the recipient of the Dean's Undergraduate Research Fellowship at New York University. A.C.-J. was the joint AMNH-NYBG Postdoctoral Fellow funded by the Cullman Program in Molecular Systematics at AMNH and NYBG.
References
- Brosnan JT, Brosnan ME. 2006. Branched-chain amino acid: Enzyme and substrate regulation. J Nutr. 136:207S–211S.
- Calyecac-Cortero GH, Cibrián-Tovar J, Bautista N, López-Collado J. 2004. Comportamiento de alimentación, cortejo, cópula y oviposición de Trichobaris championi barber (Coleoptera: Curculionidae). Agrociencia. 38:365–373.
- Caterino MS, Cho S, Sperling FH. 2000. The current state of insect molecular systematics: A thriving tower of babel. Annu Rev Entomol. 45:1–54.
- Cibrián Tovar J. 2008. Aislamiento, identificación y síntesis de la feromona del barrenador de las ramas, Copturus aguacatae Kissinger. Report to SAGARPA. Mexico City, 10 pp.
- Coria VM, Pescador A, López E, Lezama R, Salgado R, López M, Vidales A, Muñoz J, Eller FJ, Bartelt RJ, Shasha BS, Schuster DJ, Riley DG, Stansley PA, Mueller TF, Shuler KD, Johnson B, Davis JH, Sutherland CA. 2007. Autoecología del barrenador de ramas Copturus aguacatae Kissinger (Coleoptera: Circulionidae) del aguacate en Michoacán, México. Proceedings VI World Avocado Congress (Actas VI Congreso Mundial del Aguacate). Viña Del Mar, Chile. 12–16 Nov 2007.
- Edgar RC. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Eller FJ, Bartelt RJ, Shasha BS, Schuster DJ, Riley DG, Stansley PA, . 1994. Aggregation pheromone for the pepper weevil, Anthonomus eugenii Cano (Coleoptera: Circulionidae): Identification and field activity. J Chem Ecol. 20:1537–1555.
- Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10:564–567.
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 39:783–791.
- Hajibabaei M, deWaard JR, Ivanova NV, Ratnasingham S, Dooh RT, Kirk SL, . 2005. Critical factors for assembling a high volume of DNA barcodes. Philos Trans R Soc Ser B Biol Sci. 360:1959–1967.
- Hudson RR, Boos DD, Kaplan NL. 1992. A statistical test for detecting population subdivision. Mol Biol Evol. 9:138–151.
- IUPAC–IUB Joint Commission on Biochemical Nomenclature. 1983. Nomenclature and symbolism for amino acids and peptides. Available at: http://www.chem.qmul.ac.uk/iupac/AminoAcid.
- Lamb RL. 2006. Rent seeking in U.S. Mexican avocado trade. Cato J. 26:159–177.
- Lanave C, Preparata G, Saccone C, Serio G. 1984. A new method for calculating evolutionary substitution rates. J Mol Evol. 20:86–93.
- Librado P, Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.
- Nei M. 1987. Molecular evolutionary genetics. New York: Columbia University Press.
- Ochoa S, Santacruz H. 1999. Plagas del aguacatero en el Estado de México. Uruapan, Michoacán, Mexico: Facultad de Agrobiología Presidente Juárez UMSNH.
- Pogue MG, Simmons RB. 2008. A new species of Copitarsia (Lepidoptera: Noctuidae) from the neotropical region feeding on Asparagus and cut flowers. Ann Entomol Soc Am. 101:743–762.
- Ratnasingham S, Hebert PDN. 2007. BOLD: The barcode of life data system (http://www.barcodinglife.org). Mol Ecol Resour 7:355–364.
- Schilmann A, Lacasaña M, Blanco-Muñoz J, Aguilar-Garduño C, Salinas-Rodríguez A, Flores-Aldana M, Cebrián ME. 2009. Identifying pesticide use patterns among flower growers to assess occupational exposure to mixtures. Occup Environ Med. 67:323–329.
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. 1994. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 87:651–701.
- Stamatakis A. 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Stamatakis A, Ott M. 2008. Efficient computation of the phylogenetic likelihood function on multi-gene alignments and multi-core architectures. Philos Trans R Soc B Biol Sci. 363:3977–3984.
- Talavera CM, Padilla CM. 2003. Reconsideraciones tecnicas al ciclo biologico del barrenador de ramas del aguacate (Copturus aguacatae, Kissinger) . In: Actas del V Congreso Mundial del Aguacate. p 445–448.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.
- Téliz OD. 2000. El aguacate y su manejo integrado en Michoacán. Mexico City: Mundi Prensa México, S.A. de C.V . p 240.
- Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol. 39:306–314.