Abstract
Background and aims: Fishes are the main animal protein source for human beings and play a vital role in aquatic ecosystems and food webs. Fish identification can be challenging, especially in the tropics (due to high diversity), and this is particularly true for larval forms or fragmentary remains. DNA barcoding, which uses the 5′ region of the mitochondrial cytochrome c oxidase subunit I (COI) as a target gene, is an efficient method for standardized species-level identification for biodiversity assessment and conservation, pending the establishment of reference sequence libraries.
Materials and methods: In this study, fishes were collected from three rivers in southeastern Nigeria, identified morphologically, and imaged digitally. DNA was extracted, PCR-amplified, and the standard barcode region was bidirectionally sequenced for 363 individuals belonging to 70 species in 38 genera. All specimen provenance data and associated sequence information were recorded in the barcode of life data systems (BOLD; www.barcodinglife.org). Analytical tools on BOLD were used to assess the performance of barcoding to identify species.
Results: Using neighbor-joining distance comparison, the average genetic distance was 60-fold higher between species than within species, as pairwise genetic distance estimates averaged 10.29% among congeners and only 0.17% among conspecifics. Despite low levels of divergence within species, we observed river system-specific haplotype partitioning within eight species (11.4% of all species).
Conclusion: Our preliminary results suggest that DNA barcoding is very effective for species identification of Nigerian freshwater fishes.
Introduction
The inland fisheries in tropical Africa face threats both by stress from climate change and by overexploitation (Hughes et al. Citation1997). Species are becoming extinct and populations decline at an alarming but poorly understood rate. Many species may face extinction before they can be identified or described. This presents a problem for conservation planning and prioritization, because those species that have not been identified obviously cannot be protected effectively (Swartz et al. Citation2008). Caddy and Garibaldi (Citation2000) reported that only 65.09% of worldwide fishery captures reported to the FAO for the year 1996 was identified at species level, ranging from about 90% in temperate areas to less than 40% in tropical regions. Surveys into the accuracies of species identifications have not been reported, but a significant percentage of identifications may still be erroneous (Ward et al. Citation2009). The limitations inherent in morphology-based identification systems and the limited pool of taxonomists paved the way for the introduction of new molecular diagnostic tools for effective species identification. Hitherto, a wide variety of protein-based and DNA-based methods have been evaluated for the molecular identification of fish species in Africa (e.g. Waters and Cambray Citation1997; Wishart et al. Citation2006; Swartz et al. Citation2008). These studies, however, are not comparable for the purposes of species identification because they lack standardization (e.g. different regions of the mitochondrial genome such as cytochrome b and 16S rDNA were used). Hebert et al. (Citation2003) proposed a single gene sequence to discriminate the vast majority of animal species, using a 650-bp fragment of the 5′ end of the mitochondrial cytochrome c oxidase subunit I (COI) gene as a global bioidentification sequence for animals. This technology (DNA barcoding) relies on the observation that the ‘barcode’ sequence divergence within species is typically much lower than the divergence exhibited between species (Hebert et al. Citation2003), making it an effective marker for species identification and discovery. The startling efficiency of the method may arise from selective sweeps and the intricacies of mito-nuclear coadaptation, raising the profile of bioenergetics as a possible speciation mechanism (Lane Citation2009).
DNA barcoding has since gained global support as a rapid, accurate, cost-effective, and broadly applicable tool for species identification, particularly with respect to fishes as coordinated by the fish barcode of life (FISH-BOL; www.fishbol.org) campaign (Ward et al. Citation2009). Barcoding has also been adopted by the census of marine life project, a growing global conglomerate of 50 countries engaged in a 10-year initiative to assess and explain the diversity, distribution, and abundance of life in the ocean (O'Dor Citation2004). Although there has been criticism of both the philosophical and the practical underpinnings of DNA barcoding (e.g. Moritz and Cicero Citation2004; Fitzhugh Citation2006; Rubinoff et al. Citation2006; Song et al. Citation2008), its successful application for both species identification and discovery has been demonstrated in many studies, involving many taxonomic groups, for example birds (Hebert et al. Citation2004b), fish (Ward et al. Citation2005), fish parasites (Locke et al. Citation2010), bats (Clare et al. Citation2007), spiders (Barret and Hebert Citation2005), crustaceans (Costa et al. Citation2007), nematodes (Elsasser et al. Citation2009), earthworms (Chang et al. Citation2008), mosquitoes (Cywinska et al. Citation2006), and diverse arrays of Lepidoptera (Hebert et al. Citation2004a; Hajibabaei et al. Citation2006; Wilson Citation2010). In addition, DNA barcoding strategies are now being applied for other groups of organisms including plants (CBOL Plant Working Group Citation2009; Goa et al. Citation2010), macroalgae (Saunders Citation2005), fungi (Seifert et al. Citation2007; Stockinger et al. Citation2010), protists (Chantangsi et al. Citation2007), and bacteria (Sogin et al. Citation2006). Furthermore, DNA barcoding has gained wide application in forensic analysis to investigate cases of illegal poaching (Eaton et al. Citation2009), separation of species (Wilson-Wilde et al. Citation2010), gut content analysis in ecological studies (Smith et al. Citation2005; Berry et al. Citation2007; Clare et al. Citation2009), food product analysis and market substitution (Wong and Hanner Citation2008; Cohen et al. Citation2009), and Asian medicine trade regulation (Peppin et al. Citation2008). DNA barcoding has also been employed to validate the identity of biomaterial collections and cell lines (Lorenz et al. Citation2005; Cooper et al. Citation2007). A sufficient accumulation of DNA barcodes can also help conservation managers to identify interim priority areas for conservation efforts in the absence of species data. Currently, DNA barcode reference library records are available for more than 1 million sequences representing more than 94,000 species on the barcode of life data systems (BOLD; www.boldsystems.org; Ratnasingham and Hebert Citation2007), an informatics workbench aiding the acquisition, storage, analysis, and publication of DNA barcode records. Nearly, 10% of these records comprise marine and freshwater fish species (www.fishbol.org).
The COI divergence and species identification success based on DNA barcodes have been previously assessed for many freshwater fish species, for example in Canada (Hubert et al. Citation2008), Mexico and Guatemala (Valdez-Moreno et al. Citation2009), and Brazil (Carvalho et al. Citation2010; Pereira et al. Citation2010). Since, to date, there is no detailed knowledge about the diversity and distribution of freshwater fish species in Nigeria, the aim of this study was to determine whether DNA barcoding can be used as an effective tool to perform unambiguous species identification of freshwater fishes in this region, with a view toward establishing a DNA barcode reference library for utilization in biodiversity assessment and conservation for the entire country.
Materials and methods
Specimen collection and documentation
We sampled 366 fish specimens, representing 70 species, 38 genera, 20 families, comprising 25% of the 285 known fish species in all Nigerian freshwater systems, as described by Olaosebikan and Raji (Citation1998). Samples were collected from three different sites at three rivers (Afikpo, Anambra, and Ebonyi) in southeastern Nigeria between February 2008 and March 2010. The number of specimens per species collected at each site ranged from 1 to 15, with a mean of 3.5. All specimens were caught in the wild, morphologically identified in situ by visual inspection and taxonomically classified with standard guides (Olaosebikan and Raji Citation1998). Voucher specimens were imaged by digital scanning as described by Steinke et al. (Citation2009a) and preserved in 100% ethanol at the Applied Biology Department Museum, Ebonyi State University, Ebonyi, Nigeria. Details on collection, coordinates, and dates are publicly accessible in BOLD (www.barcodinglife.org) project ‘Barcoding Fishes of Eastern Nigeria’. A detailed specimen overview is given in Table S1 in the supplementary material.
DNA extraction
Muscle tissue (about 5 mm3) was extracted from the left side of each fish and preserved in 95% ethanol, using tools that were treated with DNA ELIMINase (Decon Laboratories, USA) before sampling each specimen. Genomic DNA was extracted from 1 to 2 mm3 tissue pieces, using the DNeasy Blood and Tissue kit (Qiagen, USA). The tissue pieces were incubated (and shaken at 300 rpm) overnight at 56°C in 180 μl tissue lysis buffer ATL and 20 μl proteinase K. Genomic DNA was subsequently extracted with a membrane-based approach on a Biomek FX liquid handling station (Beckman Coulter, USA) using AcroPrep 96 1.0 ml filter plates with 1.0 μm PALL glass fiber media (Ivanova et al. Citation2006).
PCR amplification
For this study, a 651-bp fragment of the standard COI barcode region was amplified using a mammal primer cocktail (Ivanova et al. Citation2007), appended with M13 (Messing Citation1983) tails to aid in a standard sequencing protocol (). Each PCR reaction mixture consisted of 6.25 μl of 10% trehalose, 2 μl ultrapure ddH2O, 1.25 μl of 10 × PCR buffer for Platinum Taq (Invitrogen, Inc., USA), 0.625 μl of 50 mM MgCl2, 0.125 μl of 10 μM primer cocktail (see ), 0.0625 μl of 10 mM dNTP mix, 0.06 μl Platinum Taq polymerase (Invitrogen, USA), and 2.0 μl template DNA to make a 12.5 μl total volume. PCR amplification reactions were conducted on Eppendorf Mastercycler gradient thermal cyclers (Brinkmann Instruments, USA). The thermocycling program consisted of a hot start of 94°C for 1 min; followed by five cycles of 94°C for 30 s, 50°C for 40 s, 72°C for 1 min; then 35 cycles of 94°C for 30 s, 54°C for 40 s, 72°C for 1 min; and then an extension of 72°C for 10 min and a final hold at 4°C.
Table I. PCR and sequencing primers used in this study.
DNA sequencing and sequence data analysis
PCR products were visualized on 2% agarose E-gel 96 plates (Invitrogen, USA) stained with ethidium bromide. PCR samples with a single visible band were processed further for sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit [Applied Biosystems, Inc. (ABI), USA]. Each forward or reverse cycle sequencing reaction mixture consisted of 0.25 μl BigDye (ABI), 1.875 μl of 5 × buffer (400 mM Tris–HCl, pH 9.0 and 10 mM MgCl2), 5 μl of 10% trehalose, 1.0 μl primer (10 μM; M13F or M13R), 0.875 μl ultrapure ddH2O, and 1.5 μl PCR product. The sequencing reaction thermocycling program consisted of 2 min at 96°C, followed by 30 cycles of 30 s at 96°C, 15 s at 55°C, and 4 min at 60°C, followed by a hold at 4°C. Bidirectional sequencing reactions were carried out with the M13 primers (), and the fluorescent signals were recorded on an ABI 3730 DNA Analyzer. Sequences were edited using the software Sequencher 4.8. To evaluate sequence identity, we used the basic local alignment search tool algorithm (Altschul et al. Citation1997) to search GenBank, and the BOLD ‘identification engine’ to query barcode records within BOLD. Top species matches obtained from both GenBank and BOLD for each specimen were compared with the specimens' species names. Sequence divergence was calculated using the Kimura 2-parameter (K2P) model (Kimura Citation1980). A mid-point rooted neighbor-joining (NJ) tree of K2P distances was created to provide graphic representation of the species divergence (Saitou and Nei Citation1987) as implemented in the ‘Sequence analysis module’ of BOLD. The sequences generated in this study are deposited in GenBank (accession numbers HM882696–HM883026) (see Table S1 for details).
Results and discussion
Deep divergence between species
We applied the technique of DNA barcoding to evaluate the suitability of this approach to unequivocally discriminate freshwater fish species from southeastern Nigeria (Africa). In this study, we obtained 363 sequences (all >500 bp) belonging to 70 species, 38 genera, and 20 families. Only three (0.8%) out of the 366 samples analyzed failed to yield a DNA barcode. No pseudogenes (short sequences with stop codons) or contaminant sequences (e.g. from bacteria) were detected by the automatic amino acid translation or contaminant screening tools when sequences were uploaded to BOLD. Sequence assemblies, electropherogram trace files, and specific primer combinations used to amplify sequenced PCR products are available for each specimen within the ‘Barcoding Fishes of Southeastern Nigeria’ project, which is publicly accessible in BOLD. A detailed specimen overview is given in Table S1.
A full K2P model-based NJ phenogram (Figure S1 in the supplementary material) shows the genetic distance between all specimens that generated a DNA barcode as described above. Our DNA barcoding approach separated all 70 species, with 10.29% distance among individuals of congeneric species (). This result is summarized in a K2P NJ tree () to provide an overview of sequence divergences between all species tested in this study. For species within higher taxonomic ranks such as families, orders, and classes, pairwise genetic divergences increased to 17.20, 21.60, and 25.05%, respectively ( and ). Some taxa, however, showed deeper genetic divergence than others. For example, the average of within-genus divergence of the silver catfish (Chrysichthys) was 11.31%, considerably larger than 8.28% of the genus Synodontis, which is itself considerably larger than the within-Alestes divergence of 4.28% (Figure S1). These differences among genera probably reflect the duration and history of species divergence, although some species within genera may be older than others (Ward et al. Citation2005).
Table II. Summary of genetic distance (according to K2P model) for increasing taxonomic levels.
Figure 1. NJ distance tree with 70 freshwater fish species (taxonomic families outside tree) of southeastern Nigeria, using K2P model distance calculation.
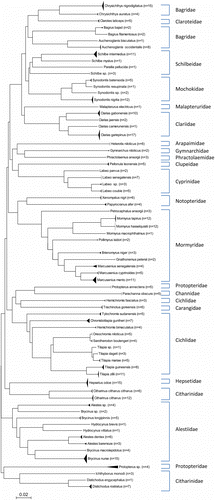
Figure 2. Genetic variability (K2P model distance of COI sequences) within different taxonomic categories. Panel A, species; panel B, genus; panel C, family; panel D, order and panel E, class.
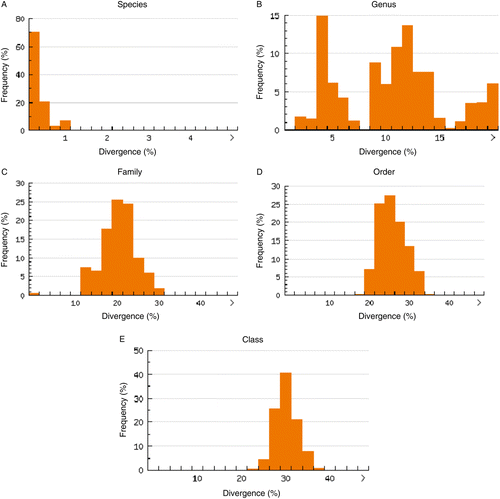
The analysis of the nearest neighbor distance (NND), which is the genetic distance between a species and its closest congeneric relative, revealed that while only 2.9% of the NND was lower than 1% (), the divergence between conspecific individuals was lower than 1% in all cases (). Congeneric NND averaged 10.9% (), which was 64-fold higher than the mean within-species distance of 0.17% and 26-fold higher than the maximum intraspecific distance of 0.42%.
Table III. Summary of southeastern Nigeria fish diversity and distribution of the genetic distance to the nearest neighbor; COI sequences of 70 species analyzed with K2P model.
Figure 3. Distribution of the genetic variability (K2P model distance of COI sequences) for the 363 individuals and 70 species analyzed. Panel A, intraspecific distance and panel B, genetic distance to the nearest neighbor. SE, standard error.
A close inspection of the K2P NJ tree () shows that distinction between two cichlids, Oreochromis niloticus and Sarotherodon boulengeri, might be ambiguous and may suggest shared haplotypes, because the genetic distance between both is only 0.46% (compare ). Such cases of shared haplotypes or low inter-specific variation may be due to introgressive hybridization, incomplete lineage sorting, taxonomic over-splitting, recent radiation, or possibly misidentification of the original specimens (Won et al. Citation2005; Hubert et al. Citation2008; Steinke et al. Citation2009b). We suspect the latter, given the fact that different genera are involved and a detailed analysis into the nature of this anomaly is underway. Berra (Citation2001) observed that the identification of cichlids might pose a taxonomic challenge with over 1000 species present in the tropics. However, the other cichlid species in this study were clearly separated (), indicating that DNA barcodes can significantly advance our understanding of the diversity in this very important group in Nigeria.
Minimal divergence but phylogeographic variation within species
The average K2P distance estimate of intraspecies divergence in this study was 0.17% (maximum 1.08%) (see ), which is lower than the previous fish DNA barcoding studies, which have reported mean conspecific divergences of 0.27% (range 0–1.95%) for 1035 salmon and trout specimens from North America (Rasmussen et al. Citation2009), 0.30% (range 0–7.42%) for 194 Canadian fish species (Hubert et al. Citation2008), 0.39% (range 0–14.08%) for 207 Australian fish species (Ward et al. Citation2005), and 0.99% (0.19% when possible misidentifications were omitted) for 72 commercial fish species in the USA (Yancy et al. Citation2008). In summary, even with a significant overlap of the genetic distances at the genus level and above (see ), the average genetic distance among congeneric species is nearly 60-fold higher than that found within species, a ratio higher than the 12-fold and 27-fold previously reported for freshwater fishes in Mexico/Guatemala (Valdez-Moreno et al. Citation2009) and Canada (Hubert et al. Citation2008), respectively. The ratio observed in this study is also much higher than the 25-fold and 26-fold reported for marine salmon and trout (Rasmussen et al. Citation2009) and ornamental marine fishes (Steinke et al. Citation2009b), respectively.
Although the primary objective of DNA barcoding is to identify species, phylogeographic structure among COI sequences within species became evident in this study. In the family Characidae, 15 individuals of Brycinus nurse were analyzed, and the full K2P NJ distance tree (Figure S1) shows two closely related (genetic distance 0.8%) but distinct clusters, with specimens BNF 178, BNF 179, BNF 181, and BNF 182 caught in Anambra river in one cluster (red color in Figure S1), while the others (BFN 102, BNF 190, BNF 191, BNF 192, BNF 194, BNF 195, BNF 196, BNF 197, BNF 198, BNF 199, and BNF 200) in the second cluster were caught in two separate but closely located rivers (Ebonyi river, blue color in Figure S1, and Afikpo river, pink color in Figure S1). A similar observation was made among the 15 specimens of Hepsetus odoe studied. The intraspecific genetic divergence was 0.20%, but there are two closely related distinct clusters of seven specimens from Anambra river (BNF 313, BNF 314, BNF 315, BNF 316, BNF 317, BNF 318, and BNF 319, red color in Figure S1) and eight specimens from Ebonyi river (BNF 320, BNF 321, BNF 322, BNF 323, BNF 324, BNF 325, BNF 326, and BNF 327, blue color in Figure S1). According to the river system from which the fish were captured, distinct clusters were also observed within six other species: Marcusenius mento, Schilbe intermedius, Clarias gabonensis, Chrysichthys nigrodigitatus, Chromidotilapia guntheri, and Citharinus citharus. These phylogeographic associations (Waters and Cambray Citation1997) may reflect a so-called ecosystem-dependent adaptive radiation (Ernst et al. Citation2003) and highlight the fact that barcoding can extend beyond just simple species identification to include phylogeographic ‘source tracking’ in many cases.
Conclusions
The results obtained in this study validate the efficacy of COI DNA barcodes for identification of freshwater fish species and will serve as a framework for future analysis of fish population structure and other applied studies such as forensic research, market analysis, food web analysis, and identification of freshwater fish eggs, larvae, fillets, and fins. Our results will also pave the way for advanced biodiversity research in Nigeria.
Supplementary Material
Download MS Word (472 KB)Acknowledgements
The authors would like to thank Rick Turner, Jaclyn McCormick, Joao Lima, John James Wilson, and Natasha Serrao (University of Guelph) for aid in imaging, DNA analysis, and data evaluation. A travel bursary was provided to CDN from the Consortium for the Barcode of Life (CBOL) to analyze specimens at the Biodiversity Institute of Ontario and the Department of Integrative Biology at the University of Guelph. Sequencing costs were paid for by the International Barcode of Life Project, with funds provided by Genome Canada (through the Ontario Genomic Institute). The authors thank the authority of Ebonyi State University PMB 053 Ebonyi (Nigeria) for the financial support of this research project.
Declarations of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. SB acknowledges support from the Canadian Barcode of Life Network provided by the Natural Sciences and Engineering Research Council of Canada.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402.
- Barret RDH, Hebert PDN. 2005. Identifying spiders through DNA barcodes. Can J Zool. 83:481–491.
- Berra TM. 2001. Freshwater fish distribution. New York, NY: Academic Press.
- Berry O, Sarre SD, Farrington L, Aitken N. 2007. Faecal DNA detection of invasive species: The case of feral foxes in Tasmania. Wildlife Res. 34:1–7.
- Caddy J, Garibaldi L. 2000. Apparent changes in the trophic composition of wild marine harvest: The perspective from FAO capture database. Ocean Coast Manag. 43:615–655.
- Carvalho DC, Oliveira DAA, Pompeu PS, Leal CG, Oliveira C, Hanner R. Deep barcode divergence in Brazilian freshwater fishes – the case of the São Francisco river basin. Mitochond DNA. 2010 (in press).
- CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proc Natl Acad Sci USA. 106:12794–12797.
- Chang CH, Rougerie R, Chen JH. 2008. Identifying earthworms through DNA barcodes: Pitfalls and promise. Pedobiologia. 52:171–180.
- Chantangsi C, Lynn DH, Brand MT, Cole JC, Hetrick N, Ikonomi P. 2007. Barcoding ciliates: A comprehensive study of 75 isolates of the genus Tetrahymena. Int J Syst Evol Microbiol. 57:2412–2425.
- Clare LE, Lim BK, Engtrom MD, Ege JL, Hebert PDN. 2007. DNA barcoding of neotropical bats: Species identification and discovery within Guyana. Mol Ecol Notes. 7:184–190.
- Clare EL, Fraser EE, Braid HE, Fenton MB, Hebert PDN. 2009. Species on the menu of a generalist predator, the eastern red bat (Lasiurus borealis): Using a molecular approach to detect arthropod prey. Mol Ecol. 18:2532–2542.
- Cohen NJ, Deeds JR, Wong ES, Hanner RH, Yancy HF, White KD, Thompson TM, Wahl M, Pham TD, Guichard FM, Huh I, Austin C, Dizikes G, Gerber SI. 2009. Public health response to puffer fish (tetrodotoxin) poisoning from mislabeled product. J Food Prot. 72:810–817.
- Cooper JK, Sykes G, King S, Cottrill K, Ivanova NV, Hanner R, Ikonomi P. 2007. Species identification in cell culture: A two-pronged molecular approach. In Vitro Cell Dev Biol Animal. 43:344–351.
- Costa FO, deWaard JR, Boutillier J, Ratnasignham S, Dooh RT, Hajibabaei M, Hebert PDN. 2007. Biological identifications through DNA barcodes: The case of the Crustacea. Can J Fish Aquat Sci. 64:272–295.
- Cywinska A, Hunter FF, Hebert PDN. 2006. Identifying Canadian mosquito species through DNA barcodes. Med Vert Entomol. 20:413–424.
- Eaton MJ, Meyers GL, Kolokotronis SO, Leslie MS, Martin AP, Amato G. 2009. Barcoding bushmeat: Molecular identification of Central African and South American harvested vertebrates. Conserv Genet. 11:1389–1404.
- Elsasser SC, Floyd R, Hebert PDN, Schulte-Hostedde AI. 2009. Species identification of North American guinea worms (Nematoda: Dracunculus) with DNA barcoding. Mol Ecol Res. 9:707–712.
- Ernst A, Becker S, Wollenzien UIA, Postius C. 2003. Ecosystem-dependent adaptive radiation of picocyanobacteria inferred from 16S rRNA and ITS-1 sequences analysis. Microbiol SGM. 149:217–228.
- Fitzhugh K. 2006. DNA barcoding: An instance of technology-driven science?. Bioscience. 56:462–463.
- Goa T, Yao H, Song J, Liu C, Zhu Y, Ma X, Pang X, Xu H, Chen S. 2010. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J Ethnopharmacol. 130:116–121.
- Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. 2006. DNA barcodes distinguish species of tropical Lepidoptera. Proc Natl Acad Sci USA. 103:968–971.
- Hebert PDN, Cywinska A, Ball S, deWard J. 2003. Biological identifications through DNA barcodes. Philos Trans R Soc Lond Biol Sci. 270:313–321.
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Halluawachss W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgeratux. Proc Natl Acad Sci USA. 2004a; 101:14812–14817.
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004b; 2:1657–1663.
- Hubert N, Hanner R, Holm E, Mandrak NE, Taylor E, Burridge M, Watkinson D, Dumont P, Curry A, Bentzen P, Zhang J, April J, Bernatchez L. 2008. Identifying Canadian freshwater fishes through DNA barcodes. PLoS One. 3:e2490.
- Hughes JB, Daily GC, Ehrlich PR. 1997. Population diversity: Its extent and extinction. Science. 278:689–692.
- Ivanova NV, deWaard JR, Hebert PDN. 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes. 6:998–1002.
- Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. 2007. Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes. 7:544–548.
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Lane N. 2009. On the origin of bar codes. Nature. 426:272–274.
- Locke SA, Mclaughlin JD, Macroglose DJ. 2010. DNA barcodes show cryptic diversity and a potential physiological basis for host specificity among Diplostomoidea (Platyhelminthes: Digenea) parasitizing freshwater fish in the St Lawrence River canal. Mol Ecol. 19:2813–2827.
- Lorenz JG, Jackson WE, Beck JC, Hanner R. 2005. The problems and promise of DNA barcodes for species diagnosis of primate biomaterials. Philos Trans R Soc Lond B Biol Sci. 360:1869–1877.
- Messing J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20–78.
- Moritz C, Cicero C. 2004. DNA barcoding: Promise and pitfalls. PLoS Biol. 2:1529–1531.
- O'Dor R. 2004. A census of marine life. Bioscience. 54:92–93.
- Olaosebikan BD, Raji A. 1998. Field guide to Nigerian freshwater fishes. New Bussa, Nigeria: Federal College of Freshwater Fisheries Technology.
- Peppin L, McEwring R, Carvalho GR, Ogden RA. 2008. A DNA based approach for the forensic identification of Asiatic black bear (Ursus thibetanus) in traditional Asian medicine. J Forensic Sci. 53:1358–1362.
- Pereira LHG, Maia GMG, Hanner R, Foresti F, Oliveira C. DNA barcodes discriminate freshwater fishes from the Paraíba do Sul River Basin, São Paulo, Brazil. Mitochond DNA. 2010.
- Rasmussen RSS, Morrissey MT, Hebert PDN. 2009. DNA barcoding of commercial important salmon and trout species (Oncorhynchus and Salmo) from North America. J Agric Food Chem. 57:8379–8385.
- Ratnasingham S, Hebert PDN. 2007. BOLD: The barcode of life data system (www.barcoding.life.org). Mol Ecol Notes. 7:355–364.
- Rubinoff D, Cameron S, Will K. 2006. A genomic perspective on the shortcomings of mitochondrial DNA for ‘barcoding’ identification. J Hered. 97:581–594.
- Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing evolutionary trees. Mol Biol Evol. 4:406–425.
- Saunders G. 2005. Applying DNA barcoding to red macroalgae: Preliminary appraisal holds promise for future applications. Philos Trans R Soc Lond B Biol Sci. 360:1879–1888.
- Seifert KA, Sampson RA, deWard JR, Houbraken J, Levesque CA, Moncalvo JM, . 2007. Prospects for fungus identification using COI DNA barcodes with Penicillium as a test case. Proc Natl Acad Sci USA. 104:3901–3906.
- Smith PJ, McVeagh SM, Allain V, Sanchez C. 2005. DNA identification of gut contents of large pelagic fishes. J Fish Biol. 67:1178–1183.
- Sogin ML, Morrison HG, Hube JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the unexpected ‘rare biosphere’. Proc Natl Acad Sci USA. 103:12115–12120.
- Song H, Buhay JE, Whiting MF. 2008. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA. 105:13486–13491.
- Steinke D, Hanner R, Hebert PDN. Rapid high-quality imaging of fishes using a flat-bed scanner. Ichthyol Res. 2009a; 56:210–211.
- Steinke D, Zemlak TS, Hebert PDN. Barcoding Nemo: DNA-based identifications for the ornamental fish trade. PLoS One. 2009b; 4:e6300.
- Stockinger H, Kruger M, Schussler A. 2010. DNA barcoding of arbuscular mycorrhizal fungi. New Phytol. 187:461–474.
- Swartz ER, Mwale M, Hanner R. 2008. A role for barcoding in the study of African fish diversity. South Afr J Sci. 104:293–298.
- Valdez-Moreno M, Ivanova NV, Elias-Gutierrez M, Contreras-Balderas S, Hebert PDN. 2009. Probing diversity in freshwater fishes from Mexico and Guatemala with DNA barcodes. J Fish Biol. 74:377–402.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. Barcoding Australia's fish species. Philos Trans R Soc Lond B Biol Sci. 360:1847–1857.
- Ward RD, Hanner R, Hebert PDN. 2009. The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol. 74:329–356.
- Waters JM, Cambray JA. 1997. Intraspecific phylogeography of the Cape galaxias from South Africa: Evidence from mitochondrial DNA sequences. J Fish Biol. 50:1329–1338.
- Wilson JJ. 2010. Assessing the value of DNA barcodes and other priority gene regions for molecular phylogenetics of Lepidoptera. PLoS ONE. 5:e10525.
- Wilson-Wilde L, Norman J, Robertson J, Sarre S, Georges A. Current issues in species identification for forensic science and the validity of using the cytochrome oxidase 1 (COI) gene. Forensic Sci Med Pathol. 2010 Doi: 10.1007/s12024-010-9172-y.
- Wishart M, Hughes J, Stewart B, Impson D. 2006. An extreme level of intra-specific divergence among Cape Peninsula populations of the Cape galaxias, Galaxias zebratus Castelnau 1861, reveals a possible species complex. Afr J Aquat Sci. 31:99–106.
- Won WJ, Sivasundar A, Wang Y, Hey J. 2005. On the origin of Lake Malawi cichlid species. A population genetic analysis of divergence. Proc Natl Acad Sci USA. 102:6581–6586.
- Wong E, Hanner RH. 2008. DNA barcoding detects market substitution in North American sea food. Food Res Int. 41:828–837.
- Yancy HF, Zemlak TS, Mason JA, Washington JD, Tenge BJ, Nguyen NT, Barnett JD, Savary WE, Hill WE, Moore MM, Fry FS, Randolph SC, Rogers PL, Hebert PDN. 2008. Potential use of DNA barcodes in regulatory science: Applications of the regulatory fish Encyclopedia. J Food Prot. 71:210–217.
