Abstract
The Fish barcode of life (FISH-BOL) initiative seeks to establish a reference sequence library of short, standardized mitochondrial gene sequences derived from the 5′ end of the cytochrome c oxidase subunit I gene (DNA barcodes) to facilitate the rapid, accurate, and cost-effective DNA-based identification of all fishes, regardless of life-stage, sex, or specimen condition. This task requires the participation of scientists from around the world and its success is predicated on the development and acceptance of standard protocols for the collection of specimens associated provenance data. Here, we provide guidelines for specimen collection, imaging, preservation, and archival, as well as meta-data collection and submission protocols developed for the FISH-BOL campaign in order to promote efficient participation in FISH-BOL by a broadening array of international participants.
Keywords::
Introduction
The Fish barcode of life initiative (FISH-BOL) (Ward et al. Citation2009) is a global effort to aid assembly of a standardized reference sequence library for all fish species, one that is derived from voucher specimens with authoritative taxonomic identifications. The benefits of barcoding fishes include facilitating species identification for all potential users, including taxonomists; highlighting specimens that represent a range expansion of known species; flagging previously unrecognized species; and, perhaps most importantly, enabling identifications where traditional methods are not applicable.
The present article outlines protocols on specimen documentation, specimen imaging, and laboratory protocols of the international campaign to barcode all fishes of the world (www.fishbol.org).
The primary work of the FISH-BOL campaign is led by various working groups that have responsibility for overseeing collections, identifications, and barcoding of the fish faunas in their region. The existing species lists associated with 19 marine and seven inland Food and Agriculture Organization of the United Nations (FAO) statistical areas provide an organizational framework for these regional teams with an initial goal of sampling five specimens from each species across each area. For certain species exhibiting broad geographic distributions, perhaps as many as 25 specimens will be sequenced under this scenario.
The FISH-BOL campaign has adopted FishBase (www.fishbase.org; CitationFroese and Pauly 2010) as the current global taxonomic authority file. In addition, we are collaborating with catalog of fishes, Integrated Taxonomic Information System (ITIS), and FishBase to resolve an integrated checklist incorporating information from each of these sources. A master list of species is available from FISH-BOL, with distributions broken down by Regional Working Group. To aid regional sampling activities, downloadable PDF or Excel sheets can be obtained for each region that tell what species were barcoded and how many barcodes exist for a particular species, as well as what species have yet to be barcoded. The objective of providing these summaries is to forestall widespread duplications of effort, including superfluous and irresponsible extractions of specimens from the environment, and unnecessary investments of time and limited resources.
FISH-BOL uses the BOL Database (BOLD) as a workbench for assembling individual projects (Ratnasingham and Hebert Citation2007). In order to aid taxonomical activities and to facilitate collaboration, BOLD offers a publicly available taxonomy browser (http://www.co1bank.uoguelph.ca/views/taxbrowser_root.php). The following protocols are written to support the incorporation of barcoding and BOLD into other existing workflows as well as harmonizing and standardizing global efforts. The present document also aims to assist collaborators in developing a participation in FISH-BOL. Please contact the chairs of your region to join (see www.fishbol.org).
Specimen documentation
Importance
Because the aim of FISH-BOL is to provide reference sequences from expert-identified voucher specimens, the act of collecting should always be accompanied by thorough documentation. This can take many forms, and not every form of documentation is desirable or even possible for each collecting event.
As collections move toward standardization for integrated information retrieval, collectors must be aware of current trends in data collection associated with biodiversity collections and must strive to obtain as much relevant data as possible in association with their collections. Modern bioinformatics initiatives can link tissue and specimen collection records with bibliographic citations, geospatial referencing information, and sequence data. FishBOL is bound to the Barcode Data Standard (CitationHanner 2009), which seeks to document barcode voucher specimens using the Darwin core triplet (institution/collection/catalog number). The Barcode Data standard defines a minimum set of information that is required to deposit a “BARCODE” annotated sequence on GenBank. BOLD supports this standard and provides fields for all required elements to GenBank such as the Darwin Core triplet, sequences, associated traces as well as primers used to retrieve the sequences as well as automated submission for the latter three.
The following additional data elements (field names in italics) are recommended for BOLD (specific details can be found online: http://www.boldsystems.org/docs/handbook.php?page = datasubprotocol):
| • | Collectors. | ||||
| • | Collection date (dd-mmm-yy). | ||||
| • | Locality: latitude and longitude using the World Geodetic System 1984, and coordinates are in degree decimal-degree format (e.g. 72.098–114.84), and the FAO region (recorded as a structured comment in the Extra Info field, see below). | ||||
| • | Elevation/depth in meters. | ||||
| • | If possible, collection gear (sampling method/effort), vessel (Notes field). | ||||
| • | Notes on habitat, microhabitat, and associations (Notes field). | ||||
| • | Sex of specimen. | ||||
| • | Life stage (adult, juvenile). | ||||
| • | Comprehensive information on the institution where specimens are vouchered and accession/catalog number (Museum voucher ID). | ||||
| • | ID. | ||||
| • | Identifier. | ||||
| • | Type status. | ||||
Identification
Error rates in museum collections and catalogs can be significant. Records in BOLD include an Identifier field and, wherever possible, the name of the person associated with a given identification should be captured. To be thorough, the taxon concept used by the identifier should also be recorded if available (i.e. original description, field guide, etc.). The Notes field provided by BOLD can host this and other additional information.
It is essential to add the appropriate FAO area into the Extra Info field of the specimen data records. This facilitates a prevailing breakdown of the barcoding progress for each working group utilizing the FISH-BOL web site.
Taxonomy
If the species is unknown, please type in genus only (in the Genus field) and leave the Species field blank; If the species is not in FishBase/Catalog of fishes but is a valid name accepted by alternative taxonomic authorities/checklists, you may wish to include it as it is (along with the authority/reference for that name); however, this will have to be discussed with the BOLD campaign manager, in order to maintain uniform naming throughout BOLD and the community. An alternative would be to type the name in question in the Extra Info field—a field that is available to all subsequent analytical functions on BOLD (e.g. it can be plotted on genetic distance-based phenograms generated by BOLD).
If a species does not have a valid name (e.g. under description or provisional morphospecies), the species epithet, ideally, should be composed of “sp.” and an alphabetic or numeric index. An example: “Sebastes sp. A”. For tentative IDs please use “cf.” ( = conformis) between genus and species epithets; for example, “Sebastes cf. alutus”.
Please note that (aside from the cases specified above) the Species field should contain a full binomen or trinomen, if applicable. Additionally, if the barcode is generated from a type specimen (or holotype, neotype, paratype, etc.) or represents topo-typic material, this information should also be recorded.
Identification levels
Entries will be identified to one of five levels of reliability depending on the taxonomic expertise of the identifier involved and their intentions following that set forth at Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia. A general definition of these levels follows:
| • | Level 1: highly reliable identification—specimen identified by (1) an internationally recognized authority of the group, or (2) a specialist that is presently studying or has reviewed the group in the region in question. | ||||
| • | Level 2: identification made with high degree of confidence at all levels—specimen identified by a trained identifier who had prior knowledge of the group in the region or used available literature to identify the specimen. | ||||
| • | Level 3: identification made with high confidence to genus but less so to species—specimen identified by (1) a trained identifier who was confident of its generic placement but did not substantiate their species identification using the literature, or (2) a trained identifier who used the literature, but still could not make a positive identification to species, or (3) an untrained identifier who used most of the available literature to make the identification. | ||||
| • | Level 4: identification made with limited confidence—specimen identified by (1) a trained identifier who was confident of its family placement, but unsure of generic or species identifications (no literature used apart from illustrations), or (2) an untrained identifier who had/used limited literature to make the identification. | ||||
| • | Level 5: identification superficial—specimen identified by (1) a trained identifier who is uncertain of the family placement of the species (cataloging identification only), (2) an untrained identifier using, at best, figures in a guide, or (3) where the status and expertise of the identifier is unknown. | ||||
Please annotate in the identification status as outlined above using the Extra Info field with an “ID-L#” preceding the FAO area designation (comma separated—e.g. ID-L3, FAO-11) such that the ID-L# can be selected to appear on BOLD generated trees. This information is absolutely crucial for curating the database as growing numbers of records become available from different sources or different geographic regions. In case an unknown specimen was identified using only the BOLD ID engine or another genetic database (e.g. GenBank), this should be mentioned in the Identifier field as “BOLD ID Engine” or “GenBank”.
Specimen imaging
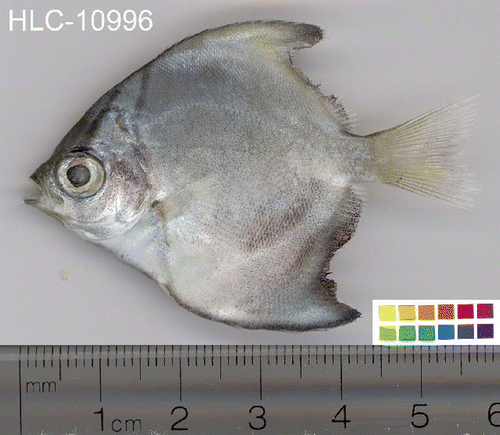
BOLD and FISH-BOL encourage image submission. This can be helpful in sorting out misidentifications and should be included whenever possible, particularly for large or small specimens that are not easily vouchered morphologically. Because bright colors fade rapidly after death, photographs should be taken as soon as possible after collection and prior to fixation if possible. Lateral photographs of the left side are most useful for taxonomic purposes. While imaging dorsal, lateral, and ventral views are critical, knowledge of the diagnostic features of a particular taxon facilitate capture of additional, more detailed photographs that are often critical for identification purposes. For example, for some specimens, close-up views of the head are also useful.
Appropriate prerequisites for a digital image associated with a DNA barcode:
| • | by convention, the left sides of fish specimens are photographed or scanned (Steinke et al. Citation2009); | ||||
| • | image format 4 × 3 to ensure optimal representation in most databases (640 × 480 pixels). The initial resolution can be higher (600–1200 dpi) to provide a good source for additional, more detailed views. Images submitted to BOLD will be automatically reduced to screen resolution. Submission of high-resolution images is encouraged; | ||||
| • | references to scale (ruler) and color (color bar or color wheel) are required in every image; and | ||||
| • | save the high-quality images of the specimen in.jpg format. | ||||
The high-quality images (see example in ) of the specimen should be submitted to BOLD in a package (compressed file, e.g. zip format) consisting of all image files and a spreadsheet with the file names and ancillary data (see BOLD online documentation: http://www.co1bank.uoguelph.ca/docs/handbook.php?page = imagesubprotocol).
DNA sampling
The current best practice in genetic resource collection involves a system of redundancy: in an ideal situation, two archival quality tissue samples will be immediately collected from each specimen; one frozen to preserve the broadest array of molecular characters possible, and one placed into a preservation fluid, such as EtOH, to serve as a back-up in case of a meltdown or loss of the frozen specimen. Several tissues are suitable for DNA extraction from fishes. These include the following.
| • | Musculature: remove one or more cubes (5–7 mm) of lateral muscle from the right side of the specimen. | ||||
| • | Gill tissue: remove one or more gill arches with attached filaments from the right side. | ||||
| • | Eye: remove the right eye from extremely small specimens such as larvae. | ||||
| • | For species with small body size, entire specimens can be placed in preservative in lieu of sub-sampling. This should be avoided unless a series of conspecifics are available for fixation in formalin for standard morphological analysis. | ||||
Tissue samples for DNA extraction should be frozen or preserved in fresh 95% EtOH and stored in a cool place, preferably in a freezer. Large pieces of tissue should be cut into small pieces ( < 5–7 mm) to permit adequate fluid penetration.
Concerns exist about whether EtOH-preserved collections more than 10–20 years old have suffered DNA degradation. Freezing EtOH-preserved tissues might minimize this problem. However, alcohol varies in several ways (e.g. hydration levels, possible contaminants). The volume of EtOH to specimen is also an important consideration, with a threefold or higher relative volume of EtOH to tissue desirable. Ultimately, EtOH is flammable and difficult to transport. Other preservatives are DNA-friendly such as RNA Later (Ambion, Austin, TX, USA), lysis buffer (Suetin et al. Citation1991), and FTA® databasing paper (Smith and Burgoyne Citation2004).
Surplus DNA extracts from barcoding could be archived at participating sequencing facilities using this platform to voucher the sequence run. However, this does not overcome the need for archiving morphological voucher specimens or tissue samples required for further comparative genetic analysis to confirm the authenticity of a suspect DNA extract and/or associated barcode sequence.
DNA barcoding laboratory protocols
DNA extraction
Simple methods of DNA extraction, such as a proteinase K digestion and/or Chelex extraction, are usually sufficient, especially while dealing with fresh specimens, but often give poor quality DNA extracts. Various commercial kits are also available and were used with good success, although they tend to be expensive when conducting high-throughput work. We recommend a standard protocol—a glass-fiber-based system (Ivanova et al. Citation2006) that works very efficiently for samples of various ages.
DNA amplification and sequencing
A standard 12.5μl Polymerase Chain Reaction (PCR) mix includes 6.25μl of 10% trehalose, 2.00μl ultrapure water, 1.25μl of 10 × PCR buffer (200 mM Tris–HCl, pH 8.4, 500 mM KCl), 0.625μl MgCl2 (50 mM), 0.125μl each primer (0.01 mM; see ), 0.062μl each dNTP (10 mM), 0.060μl Platinum® Taq Polymerase (Invitrogen, Carlsbad, CA, USA), and 2.0μl DNA template.
Table I. PCR and sequencing primers.
A 650 bp barcode region of Cytochrome C oxidase Subunit I (COI) can be subsequently amplified under the following thermal conditions: 2 min at 95°C; 35 cycles of 0.5 min at 94°C, 0.5 min at 52°C, and 1 min at 72°C; 10 min at 72°C; held at 4°C.
PCR products are visualized on 1.2% agarose gels and visible products are selected for sequencing. However, as next-generation PCR machines come of age, gel electrophoresis will probably be replaced by high-resolution melt analysis, which has the advantage of being able to differentiate two competing amplicons of approximately equal size (Vossen et al. Citation2009). This is noteworthy because multiple independent amplicons can arise within a single PCR because of nonspecific amplification and yet go undetected by typical gel electrophoresis. Successful PCR products can be bi-directionally sequenced using PCR primer pairs (Ward et al. Citation2005) or the sequencing primers M13F or M13R for multiplex reactions (Ivanova et al. Citation2007). Reaction clean-up is advisable prior to sequencing, but can be omitted if PCR conditions are well optimized.
Data upload
Sequence information can be uploaded in BOLD for several specimens at once in FASTA format (see BOLD online documentation: http://www.co1bank.uoguelph.ca/docs/handbook.php?page = seqsubprotocol). The FASTA header line must conform to the following format: it should begin “>” followed by the Process ID or Sample ID, followed by a bar (“|”), followed by any other information the user wishes to add. There can be no spaces before the end of the Process ID or Sample ID. Trace files can be uploaded in.ab1 or.scf format and corresponding phred scores in.phd format using a routine similar to image submission (see BOLD online documentation: http://www.co1bank.uoguelph.ca/docs/handbook.php?page = tracesubprotocol).
Acknowledgements
The authors would like to thank the founding members of FISH-BOL for their efforts to push the campaign forward. They would also like to acknowledge the many students who have aided development of these protocols at the University of Guelph, including Natalia Ivanova, Christa Maitland, Rachel Breese, and particularly Heather Braid. The FISH-BOL campaign has benefited from organizational support provided by the Consortium for the Barcode of Life as well as operating support provided by the Canadian Barcode of Life Network (with funding from Genome Canada and NSERC) and the International Barcode of Life project. D.S. was supported by funding from the Alfred P. Sloan Foundation to MarBOL.
Declarations of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Froese R, Pauly D. 2010. Fishbase. Available at: www.fishbase.org version. Accessed in July 2010.
- Hanner R. 2009. Data standards for BARCODE records in INSDC (BRIs). Available at: http://www.barcoding.si.edu/PDF/DWG_data_standards-Final.pdf.
- Ivanova NV, Dewaard JR, Hebert PDN. 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes. 6:998–1002.
- Ivanova NV, Zemlak TS, Hanner R, Hebert PDN. 2007. Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes. 7:544–548.
- Messing J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20–78.
- Ratnasingham S, Hebert P. 2007. BOLD: The barcode of life data system. Mol Ecol Notes. 7:355–364.
- Smith LM, Burgoyne LA. 2004. Collecting, archiving and processing DNA from wildlife samples using FTA databasing paper. BMC Ecol. 4:4.
- Steinke D, Hanner R, Hebert PDN. 2009. Rapid high-quality imaging of fishes using a flat-bed scanner. Ichthyol Res. 56:210–211.
- Suetin G, White BN, Boag PT. 1991. Preservation of avian blood and tissue samples for DNA analysis. Can J Zool. 69:82–90.
- Vossen RH, Aten E, Roos A, den Dunnen JT. 2009. High-resolution melting analysis (HRMA): More than just sequence variant screening. Hum Mutat. 30:860–866.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia's fish species. Philos Trans R Soc B Biol Sci. 360:1847–1857.
- Ward RD, Hanner R, Hebert PDN. 2009. The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol. 74:329–356.