Abstract
Background and aims: Here we describe preliminary efforts to integrate DNA barcoding into an ongoing inventory of the Lower Congo River (LCR) ichthyofauna. The 350 km stretch of the LCR from Pool Malebo to Boma includes the world's largest river rapids. The LCR ichthyofauna is hyperdiverse and rich in endemism due to high habitat heterogeneity, numerous dispersal barriers, and its downstream location in the basin.
Materials and methods: We have documented 328 species from the LCR, 25% of which are thought to be endemic. In addition to detailing progress made to generate a reference sequence library of DNA barcodes for these fishes, we ask how DNA can be used at the current stage of the Fish Barcode of Life initiative, as a work in progress currently of limited utility to a wide audience. Two possibilities that we explore are the potential for DNA barcodes to generate discrete diagnostic characters for species, and to help resolve problematic taxa lacking clear morphologically diagnostic characters such as many species of the cyprinid genus Labeo, which we use as a case study.
Results: Our molecular analysis helped to clarify the validity of some species that were the subject of historical debate, and we were able to construct a molecular key for all monophyletic and morphologically recognizable species. Several species sampled from across the Congo Basin and widely distributed throughout Central and West Africa were recovered as paraphyletic based on our molecular data.
Conclusion: Our study underscores the importance of generating reference barcodes for specimens collected from, or in close proximity to, type localities, particularly where species are poorly understood taxonomically and the extent of their geographical distributions have yet to be established.
Introduction
From its main source in the Katanga Highlands of southeastern Congo to its exodus into the Atlantic Ocean in western Bas Congo, the main channel of the Congo River forms a 4374 km arc twice bisecting the Equator (Runge Citation2007). The river and its tributaries drain an area the size of Europe and represent over 14,500 km of navigable passage across Central Africa, providing food, transportation, and livelihoods for the over 30 million people who live in this vast region. Over a decade ago, 780 species of fishes were recorded from the Congo Basin (Lévêque Citation1997) with 83% considered endemic to the region (Chapman and Chapman Citation2001), but despite more than a century of ichthyological investigation (Boulenger Citation1901) much of the basin remains virtually unexplored (Thieme et al. Citation2005) and species discovery is the norm when new localities are surveyed or regions revisited. Due to limited exploration throughout the region, incomplete sampling, and a chronic lack of up-to-date taxonomic and ecological knowledge, it is widely acknowledged that the total number of fish species is probably far greater than currently documented. The present study stands in contrast to previous barcoding studies of freshwater fishes that have focused on better-understood taxa with intensive taxonomic coverage, including tissue samples available for most species over a wide geographic range (e.g. freshwater fishes of Canada; Hubert et al. Citation2008).
Here, we investigate the utility of DNA barcoding fishes in a region that could be considered Canada's DNA barcoding antipode: the Lower Congo River (LCR). At the western edge of the Congo basin, near the twin capitals of Kinshasa (Democratic Republic of Congo [DRC]) and Brazzaville (Republic of Congo [RC]), the river spills over a rocky sill at the edge of Pool Malebo and plunges down a narrow gorge cut through the Crystal Mountains of the Atlantic Rise. In a short stretch of about 350 km, the LCR drops 280 m in a stepped series of rapids before it begins its outflow into the Atlantic Ocean (). The first detailed ichthyological survey of the LCR rapids (Roberts and Stewart Citation1976) recorded the presence of 147 fish species including 34 LCR fishes, suggesting that this is an extremely species-rich stretch of river. Survey efforts over the past 4 years by ichthyologists from the American Museum of Natural History (AMNH) in collaboration with Congolese researchers at the Universities of Kinshasa (DRC) and Marien Ngouabi (RC) have more than doubled these counts, although considerable taxonomic uncertainty persists for many of these putative species (). Additionally, in situ measurements have shown that river flow in much of the LCR is highly energetic, and that complex flow structure and bathymetry are probably playing a key role in the isolation of fish populations; this isolation over small geographic scales may in turn account for the extraordinarily high levels of species endemism (Markert et al. Citation2010). In light of the extreme hydrological conditions that appear to isolate many LCR fish populations from those in the remainder of the basin, it is highly likely that further investigation will reveal numerous additional endemics that are currently (erroneously) assigned to more widespread species, and cytochrome c oxidase subunit I (COI) diagnostics could be a preliminary tool for such investigation. In light of the documentation of such high levels of species richness and endemism in the LCR, an answer to the question “Why barcode the fishes of the LCR?” is easy: it provides a significant contribution to the Fish Barcode of Life (FISH-BOL) campaign to establish a barcode reference library for all the world's fishes. But beyond that, it is hoped that our experiences can help to clarify the utility and limitations of DNA barcodes as a tool for biodiversity inventory of the taxonomically less-well-documented fish faunas of tropical freshwaters.
Figure 1. Study system. Unlike most large rivers, the Congo lacks a delta, and much of the African continent's interior drains into the Atlantic through a narrow stretch of river comprising the world's greatest rapids. For 350 km, between the twin capitals of Brazzaville and Kinshasa to Boma, the Congo drops approximately 280 m in a series of 66 major cataracts. Labeo for this study were collected at localities demarcated with stars. Black, proximity of Kisangani; dark gray, Lulua River; light gray, West Africa and Lower Guinea; and white (boxed), Lower Congo rapids.
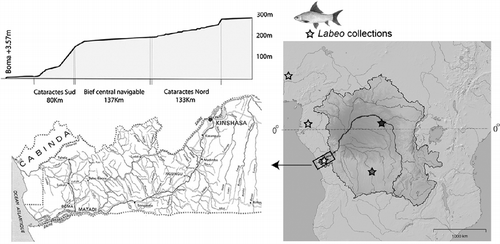
Table I. Fishes recorded from the LCR.
The consensus among participants attending the first meeting of the African Regional Working Group of FISH-BOL (PAFFA Citation2008) was that the most efficient manner to collect DNA barcodes for the African ichthyofauna was through collaboration with existing biodiversity inventories and taxonomic initiatives on the continent. Indeed, the value of an integrative approach to taxonomy is recognized (Padial et al. Citation2010) and was documented for other large-scale inventory projects involving tropical biodiversity (e.g. Janzen et al. 2008). Such integration is deemed beneficial for several reasons. First, and most pragmatically, COI sequences could usefully augment morphological and molecular phylogenetic analyses already underway on LCR fishes. Secondly, the use of COI sequence diagnostics in concert with morphological characters in species descriptions (Amato et al. Citation1999; Victor Citation2007) is recognized as an increasingly useful synergy. Incorporating gene sequences from type specimens (or retrospectively from topotypical material) optimizes the accuracy of future barcode identifications—which is particularly important when faunas are poorly understood taxonomically and geographical distributions undefined—while allowing for these data to be used as any other discrete character (DeSalle Citation2006) for unambiguously diagnosing species (DeSalle et al. Citation2005; Lowenstein et al. Citation2009).
While comprehensive reference sequences from systematics-driven initiatives can be added to the Barcode of Life Data System (BOLD; Ratnasingham and Hebert Citation2007) with relative ease and can be used to generate species hypotheses or identifications, generating complete reference libraries in localities like the LCR is a far greater challenge. Clearly accession to BOLD of COI sequences of the many LCR endemics that have striking morphologies, or have already been subject to taxonomic scrutiny, is unproblematic and can be fast-tracked to provide an authoritative reference for the molecular identification of these geographically delimited taxa. Unfortunately, such is not the case for many of the other LCR taxa, and although considerable progress was made, it is unlikely that a comprehensive DNA barcode library for the LCR will be achieved within the decade. This is primarily because insufficient taxonomic research was conducted on portions of the region's ichthyofauna to allow for unambiguous species assignment, and in many cases the species historically recorded from this river stretch have yet to be relocated. Biodiversity science—either barcode-aided or not—is hindered when there are few taxonomic authorities and insufficient past scholarship to allow unambiguous identification of specimens. Such is clearly the case for many of the poorly diagnosed taxa that were reported from the LCR (), and in many cases determining whether individuals collected in the LCR are novel to the system (localized endemics) or populations of extralimital species requires significant revisionary effort and would benefit greatly from access to sequences from type (or, more realistically, topotypical) material. At the current stage of the FISH-BOL campaign, perhaps its greatest beneficiaries may be taxonomists working on such historically contentious taxa that have yet to be subjected to molecular analysis and for which morphological diagnostics have yet to be identified. One such taxonomically challenging assemblage considered here comprises the large-bodied, heavily harvested cyprinids of the species-rich genus Labeo Cuvier, 1817, a genus that is particularly well-represented in the LCR () and that poses significant problems with respect to species delimitation and assignment ().
Table II. Specimens, sample identification codes, and collecting localities for Labeo from the LCR, Lualua River, Middle Congo (Kisangani), and Cameroon.
Monophyly of this large Afro-Asian genus is strongly supported by morphological apomorphies (Tshibwabwa Citation1997; Stiassny and Getahun Citation2007). While Tshibwabwa (Citation1997) found support for the monophyly of the African representatives, this result has yet to be assessed using molecular data. Boulenger (Citation1909) compiled the first identification key for 36 African species, and subsequently more than 100 have been described, although Reid (Citation1985) accepted only 46 as valid, writing: “In my investigation I have only rarely found a character unique to and possessed by all of the included individuals of any one ‘species’—overlapping variation is the general case” (p. 12). Although others argue that Reid's estimate is overly conservative (Roberts Citation1986; Skelton Citation1994; Tshibwabwa Citation1997; Tshibwabwa et al. Citation2006), recognition of the highly problematic alpha-taxonomy of the group resulting from a paucity of morphological diagnostics is one of few points of agreement. Reid (Citation1985) sorted the African Labeo into six species groups based on anatomy and morphometrics, but conceded that the divisions may not be monophyletic and that their relationships may lie with certain Asian Labeo. Subsequent workers have challenged the validity of Reid's divisions (Roberts Citation1986; Thys van den Audenaerde Citation1987; Tshibwabwa Citation1997). Using 17 morphological characters, however, Tshibwabwa (Citation1997) affirmed the two main morphological groups of African Labeo recognized by Reid—the “papillate group” and the “plicate group”—differentiated on the basis of the disposition of the papillae on the upper labial fold of the upper jaw and lower lip. These commercially important (Skelton et al. Citation1991; Tshibwabwa and Teugels Citation1995; Tshibwabwa Citation1997; Gordon Citation2006), yet taxonomically poorly understood cyprinids therefore represent an interesting case study for the application of DNA barcoding in a hyperdiverse tropical riverine system.
Here, we explore whether COI assists in resolving some of the taxonomic uncertainties and conflicting synonymies proposed by previous authors, and investigate whether the morphologically well-defined Labeo species can also be diagnosed using nucleotide characters. Based on these data, we present a preliminary molecular phylogeny of species relationships among the available sampling of Labeo from the Congo basin, the Lower Guinean ichthyofaunal province, several Southeast Asian Labeo and cyprinid outgroups using nuclear rag1 and mtDNA COI and control region sequences. We then compare this result with a gene tree derived from COI.
Materials and methods
Collecting localities and sampling
Large synoptic collections of fishes were made over four field seasons (2006–2009) at multiple sites along a 350-km stretch of the LCR, in Pool Malebo and from selected sites along affluent rivers and streams of the Pool and along the LCR (). Due to a combination of limited river access and extreme hydrological conditions throughout much of the river reach, sampling in main channel habitats of the LCR is extremely challenging. As a result, most collecting effort focused on relatively sheltered shoreline habitats and, despite intensive efforts, many of the species historically recorded as present in the LCR have yet to be re-collected (see below).
Collections and specimen processing was conducted in accordance with the American Fisheries Society Guidelines (Nickum Citation2004). For many of the species collected, tissue samples were field preserved in 95% ethanol, and voucher specimens were preserved in formalin and transferred to 70% ethanol for morphological analysis. Voucher specimens were accessioned into the collections of the Department of Ichthyology, AMNH, New York and the Zoologische Staatssammlung (ZSM), Munich. Specimen provenance data and sequence information (see below) were deposited on BOLD (project code: AMNHI) in accordance with FISH-BOL guidelines (Ward et al. Citation2009).
Labeo analysis
Specimens incorporated in the molecular analysis, excluding those downloaded from GenBank, were assigned to species by a world authority on Labeo taxonomy (S.M. Tshibwabwa), and the broad geographic areas from which they were collected are represented with shaded stars in . Putative species from the LCR, four of which appear to represent range extensions into the LCR (marked with an asterisk), are included: Labeo annectens* Boulenger, 1903, Labeo barbatus Boulenger, 1898, Labeo cyclorhynchus Boulenger, 1899, Labeo fulakariensis Tshibwabwa et al., Citation2006, Labeo greenii Boulenger, 1902, Labeo lineatus Boulenger, 1898, Labeo longipinnis Boulenger, 1898, Labeo lukulae* Boulenger, 1902, Labeo maleboensis Tshibwabwa, Citation1997 (from Pool Malebo and Nsele River), Labeo nasus Boulenger, 1899, Labeo parvus Boulenger, 1902, Labeo cf. quadribarbis* Poll and Gosse, 1963, Labeo simpsoni* Ricardo-Bertram, 1943, Labeo sorex Nichols and Griscom, 1917, and Labeo weeksii Boulenger, Citation1909. In addition, specimens conforming to the description of Labeo lividus Roberts and Stewart, Citation1976 (considered a junior synonym of L. barbatus) and individuals of two putative new species from LCR were also incorporated in the study. Species from the Lulua River, a large left bank tributary of the Kasai River located in the Kasai Orientale Province, south Central DRC, identified as Labeo chariensis Pellegrin, 1904, Labeo cf. coubie Rüppell, 1832, L. cyclorhynchus, L. greenii, L. lineatus, L. parvus, L. cf. parvus, Labeo cf. rectipinnis Tshbwabwa, 1997, and L. simpsoni, and from the upper Congo River from the vicinity of Kisangani species identified as L. fulakariensis, L. greenii, and L. parvus were also included. Additionally, due to tissue availability, we include the Lower Guinean Labeo batesii Boulenger, 1911, Labeo nunensis Pellegrin, 1929, and Labeo sanagaensis Tshibwabwa, Citation1997 (species identification of the three lower guinean Labeo was by M.L.J.S.) and the West African Labeo senegalensis Valenciennes, 1842. GenBank data were used for L. senegalensis (EU711151, NC_008657, NC_008657), the Asian Labeo calbasu Hamilton, 1822 (EU417758), Labeo rohita Hamilton, 1822 (EU030668), Labeo gonius Hamilton, 1822 (EU417801), the Labeonine Bangana ariza Hamilton, 1807 (EU417809), and the danionines Opsariichthys uncirostris Temminck and Schlegel, 1846 (EF452894, FJ197126, EU154953), and Zacco platypus Temminck and Schlegel, 1846 (EF452896, EF452848, AY095338).
DNA extraction
Two methods were used for DNA extraction. Manual extractions employed a DNeasy Blood and Tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions, while the majority of extractions were completed using an automated silica-based DNA extraction method. In brief, tissue was digested overnight in vertebrate lysis buffer with Proteinase K at 56°C, and genomic DNA was subsequently extracted using a membrane-based approach on a Biomek NX liquid handling station (Beckman Coulter, Brea, CA, USA) using AcroPrep 96 1 ml filter plates (PALL, Port Washington, NY, USA) with 1.0 μm PALL glass fiber media following Ivanova et al. (2006).
PCR and fragment analysis
PCR products were visualized on 2% agarose E-gel 96 plates (Invitrogen, Carlsbad, CA, USA) stained with ethidium bromide. PCR samples with a single visible band were processed further for sequencing. We amplified three markers with individual sets of primers. For the nuclear gene rag1, we used the following two primer pairs (López et al. Citation2004) to produce two overlapping fragments that allows the generation of a single contig of approximately 1500 bp: RAG1F1 (5′-CTGAGCTGCAGTCAGTACCATAAGATGT-3′) with RAG1_JHL_Ri (5′-TTCATCGTGGCTGCGTGTGA-3′), and RAG1R1 (5′-CTGAGTCCTTGTGAGCTTCCATRAAYTT-3′) with RAG1_JHL_Fi (5′-ATGCACGCTCTGCGACTCAA-3′). The RAG1_JHL internal primers were designed for this study using Primer3 (Rozen and Skaletsky Citation2000). The thermal cycling program for this fragment was: 95°C for 2 min, then 35 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min, followed by a final step at 72°C for 10 min. Part of the mitochondrial DNA control region (about 800 bp) was amplified with the following set of primers (Gilles et al. Citation2001): ESTFOR (5′-CATCGGTCTTGTAATCCGAAGAT-3′) and NewCRev (5′-GTTTCGGGGTTTGACAAGGATA-3′). The thermal program for this fragment was: 95°C for 2 min, then five cycles of 94°C for 15 s, 48°C for 45 s, and 72°C for 150 s, followed by 30 cycles at 94°C for 15 s, 52°C for 45 s, and 72°C for 150 s, finished with an elongation step of 72°C for 7 min. The mitochondrial COI gene was amplified with the fish primer cocktail and protocol described by Ivanova et al. (Citation2007).
DNA sequencing
We bi-directionally sequenced all marker genes using BigDye Terminator v3.1 chemistry (Applied Biosystems, Inc. [ABI], Carlsbad, CA, USA). Each sequencing reaction (forward or reverse) contained 1 μl of the following components: BigDye, 5 × sequencing buffer, forward or reverse primer (10 μM, see primer pairs above), and template DNA (1.5 μl for nonabundant fragments). The PCR fragments generated with the fish primer cocktail (see above) are M13-tailed, and were sequenced bi-directionally with M13 (forward and reverse) primers to yield complete 2x coverage of the barcode fragment. Water was added to a total volume of 14 μl per reaction, and subjected to a thermal cycling program in an Eppendorf Mastercycler model ep (Eppendorf, Hauppauge, NY, USA), gradient S or Pro S: 96°C for 2 min, then 30 cycles of 96°C for 30 s, 55°C for 15 s, and 60°C for 4 min. Electropherograms were generated with an ABI 3730xl sequencer, and imported in the programs CodonCode Aligner v3.0.1 (CodonCode Corp., Dedham, MA, USA) and Geneious Pro v4.9.3 (Biomatters, Auckland, New Zealand) for sequence contig assembly and manual editing.
Phylogenetic analysis
Labeo sequences were aligned using MAFFT v6 (Katoh and Toh Citation2008) and the alignments edited using Geneious. Ambiguously aligned nucleotide positions in the control region were removed prior to analysis. Sequence files for three loci (COI, 652 bp; rag1 1393 bp; control region, 788 bp) were concatenated (2833 bp) using Geneious and exported as FASTA-formatted files, which were in turn converted to a “relaxed” PHYLIP format (i.e. allowing taxon labels to occupy up to 256 characters) using a Perl script available on the webpage of A. Stamatakis (http://wwwkramer.in.tum.de/exelixis/software.html). Maximum likelihood phylogenetic analyses were performed using RAxML v7.2.3 (Stamatakis Citation2006; Stamatakis et al. Citation2008) implemented on the CIPRES Portal v2.2 (http://www.phylo.org) at the San Diego Supercomputer Center (CitationMiller et al. 2009). Separate analyses using the same specimens were conducted on the COI and the concatenated three-locus (COI+rag1 + control region) alignments. Outgroup taxa included two nonlabeonine cyprinids, Z. platypus, and O. uncirostris. Analyses included 200 inferences on the original alignment and 10,000 bootstrap replicates, using the general time-reversible substitution model (Lanave et al. Citation1984) with among-site rate heterogeneity modeled by the Γ distribution and four discrete rate categories (Yang Citation1994) for both bootstrapping and final maximum-likelihood optimization and using default parameter settings. Tree figures were prepared using FigTree v1.3.1 (CitationRambaut 2010).
COI diagnostic key
A diagnostic key was constructed using Geneious to visualize the alignment and conduct Population Aggregation Analysis (Davis and Nixon Citation1992) by eye, defining species using “characteristic attributes” (CAs; Sarkar et al. Citation2002). Species “nucleotide diagnostics” (e.g. Wong et al. Citation2009) were created using “pure CAs”, which are nucleotide characters unique to one species, and “compound CAs”, which are combinations of nucleotides at multiple sites that are diagnostic for that species.
Results
Species diversity
provides a comprehensive list of species found to date in the LCR main channel and/or its affluent rivers (). The list was compiled after the review of available taxonomic literature, examination of collections housed in major US and European museums (where possible including type specimens), consultation with taxonomic authorities, and detailed examination of many thousands of specimens collected over the past 4 years by ichthyologists from the AMNH in collaboration with Congolese researchers at the Universities of Kinshasa (DRC), and Marien Ngouabi (RC). Remarkably, given that the LCR comprises only 2% of the total area of the Congo basin (Roberts and Stewart Citation1976), we record the presence of 328 species of which 84 are currently recognized as endemic to the system. We note, however, that considerable taxonomic uncertainty persists for many putative LCR species, and identification is tentative for those indicated in with a question mark (?). This is a reflection of a lack of clear morphological diagnostics, often combined with poorly established geographic ranges for many species. It is noteworthy that 160 LCR species are represented by type specimens (wholly or in part) collected in, or in close vicinity to, the LCR (). Family-level representation in LCR is also high, and includes all but three of the families present in the entire Congo basin (Phractolaemidae, Kneriidae, and Pantodontidae). Of this diversity, we have progressed in archiving COI sequences for 110 species, 20 of which are endemic to the LCR. These are noted in the publicly accessible BOLD project (code: AMNHI) and the type status of the material examined was noted therein. Tissues from an additional 38 endemics and 112 nonendemics remain to be processed. Twenty-four endemics for which we lack tissues are recorded from the system ().
Phylogenetic analysis of Labeo
The topology of the COI tree () was in general agreement with the three-gene tree (), although with considerably less bootstrap support for most clades. The placement of L. batesii and L. annectens in the COI tree is in disagreement with the three-gene tree. When trees were constructed using rag1+COI, as well as separate rag1 and control region trees, all agreed with the multi-gene topology, suggesting that the aberrant COI result was not a case of introgression.
Figure 2. COI Labeo gene tree. Maximum-likelihood phylogram showing branch bootstrap support above the 70% (log-likelihood score of best tree: − 4137.464790). Outgroup taxa omitted. Circle, Asian Labeo; “a” and “b”, the two main clades discussed in the text. Specimen codes ending in “K” denote individuals caught in the vicinity of Kisangani (Upper Congo); “L” collected from the Lualua, a large southern tributary of the Congo River; and “C” describes specimens collected from Cameroon. One species, L. senegalensis (GenBank accession number NC008657), inhabits western Africa. The placement of L. batesii and L. annectens is in disagreement with the relationships recovered using rag1 and the control region singularly (not shown) and in concert ().
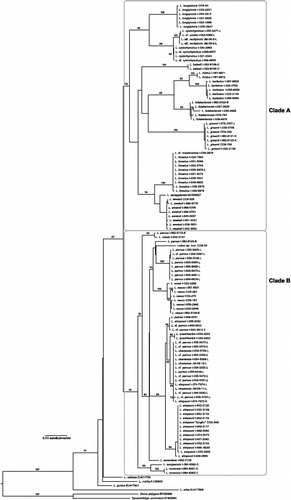
Figure 3. Phylogenetic tree of the concatenated sequence dataset (control region, COI, and rag1) of African Labeo taxa. Maximum-likelihood phylogram showing branch bootstrap support above the 70% (final GAMMA-based score of best tree: − 14613.154605). Two primary clades were recovered (A and B). The symbols denote Labeo that belong to the groupings proposed by Reid (Citation1985). Square, niloticus group; circle, macrostomus group; triangle, coubie group; diamond, forskallii group. Nine individuals represented on the COI phylogenetic tree () were omitted due to failure to amplify all genes.
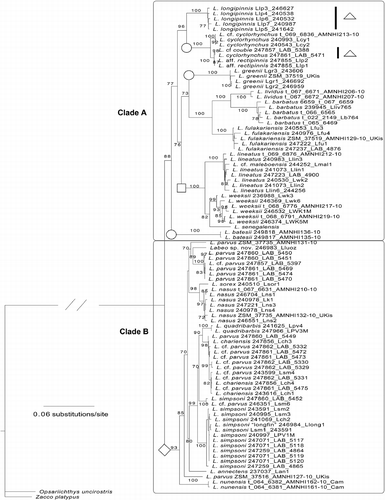
Our analysis supports a monophyletic Labeo and provides provisional support for a monophyletic African Labeo, although we note that our sampling of Asian species is minimal and that none of the southern African Labeo species were incorporated in our study. Nonetheless, it is noteworthy that some of the Asian species included have previously been suggested as being closely related to certain of the African taxa; for example, L. gonius was allied with the Labeo niloticus species group, and Labeo ariza ( = Bangana ariza) with the Labeo macrostomus species group by Reid (Citation1985). Surprisingly, only the “papillate group” is shown here to be monophyletic for the three genes examined in this work. Relationships of the “papillate clade” lay with a subset of the “plicate group”, thus rendering the “plicate group”, as broadly conceived, paraphyletic. Our analyses, while preliminary, retrieve two main clades within the African Labeo sampled (; Clades A and B). In Clade A, the sole specimen of L. maleboensis shares an identical haplotype with L. lineatus. This is noteworthy as Tshibwabwa (Citation1997) lists a series of seemingly stable morphometric and meristic features that readily differentiate L. maleboensis from L. lineatus. Our study clearly indicates that further investigation of this putative species pair with greater sampling is in order. Regardless of specific status, this group is in turn resolved as sister to a L. senegalensis+L. weeksii grouping. Sister to this “papillate clade” are a morphologically highly diversified subgroup of “plicate-lipped” species. Among these, L. greenii is similar in overall morphology to L. fulakariensis, the latter having been identified as L. greenii in museum collections prior to recognition as a distinct species (Tshibwabwa et al. Citation2006). Despite the close morphological resemblance of L. greenii and L. fulakariensis, there is strong molecular support for the alignment of the latter with the phenetically divergent L. barbatus+L. lividus clade.
Labeo lividus was described from the LRC (Roberts and Stewart Citation1976), but is considered by both Reid (Citation1985) and Tshibwabwa (Citation1997) as a junior synonym of L. barbatus. In this case, a re-examination of specimens from LCR confirms that the differentiating diagnostics proposed by Roberts and Stewart (Citation1976) for L. lividus—such as shorter, paler barbels and larger, more heavily keratinized lip papillae—do differentiate between specimens that resolve into two clades within L. barbatus sensu lato in this analysis.
Our sampling of L. cyclorhynchus is rendered paraphyletic by one Lulua individual that is allied to specimens identified as L. rectipinnis and L. cf. coubie, suggesting that this group, which is sister to a well-diagnosed L. longipinnis, is also in need of further taxonomic scrutiny. L. batesii, a Lower Guinean species, is resolved as sister to the above-mentioned taxa (but is anomalously placed as sister to the L. fulakariensis, L. lividus, L. barbatus, and L. greenii subclade in the COI tree).
Clade B () consists of the remaining plicate-lipped Congolese species sampled and the Lower Guinean L. sanagaensis and L. nunensis that are resolved as sister to them. A putative novel species L. “longfin”, collected in LCR, shared the same haplotype with 10 individuals from LCR identified as L. simpsoni and as such is probably best considered as a morphologically aberrant member of that species. While the LCR L. simpsoni (including the “longfin” form) formed a clade, species identified as L. simpsoni from the Lulua River were not phylogenetically resolved with the LCR individuals, nor did they group together in any of our trees. Two individuals from LCR identified as L. quadribarbis—a species synonomized with L. parvus by Reid (Citation1985) but shown to be morphologically distinct and considered valid by Tshibwabwa (1997)—are resolved as sister to a morphologically heterogeneous clade consisting of specimens identified as L. chariensis (n = 4), L. cf. parvus (n = 6), L. parvus (n = 1), and L. simpsoni (n = 1)—all individuals were from the Lulua and all share an identical haplotype. Despite some differences in dorsal fin shape, scale counts, and lateral line coloration used to morphologically differentiate L. chariensis, L. parvus, L. cf. parvus, and L. simpsoni, our genetic results suggest that these characters may exhibit considerable variability, and as such do not provide an adequate basis for species delimitation. In this regard, the L. parvus phenotype is particularly problematical and individuals identified as this species are represented in four different clades in our analysis. The two individuals sampled from Kisangani (Middle Congo), despite morphological similarity, are widely divergent genetically: one is most closely related to an individual collected in LCR and identified as L. lukulae, while the other is sister to an undescribed species from the LCR (L. sp. nov. CO8-53). In contrast, but expected based on morphological similarity, the L. sorex+L. nasus clade is well supported by molecular data.
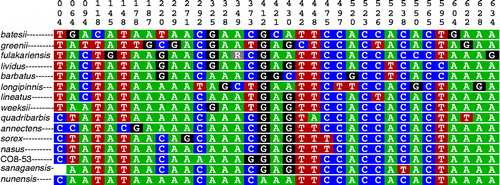
The character-based (COI) key successfully differentiates the following morphologically distinct species: L. annectens, L. barbatus, L. batesii, L. fulakariensis, L. greenii, L. longipinnis, L. quadribarbis, L. sanagaensis, L. sorex, and L. sp. nov. CO8-53 with pure CAs (). Compound CAs differentiated the remaining species: L. lividus, L. lineatus, L. nasus, and L. weeksii. Labeo fulakariensis was the only species with multiple individuals sampled (n = 5) that was polymorphic at one of the sites that was diagnostic for other taxa.
Figure 4. COI character-based key for select species of Labeo. Top vertical numbers denote positions of the 652 bp fragment analyzed. L. annectens, L. barbatus, L. batesii, L. fulakariensis, L. greenii, L. lineatus, L. longipinnis, L. nunensis, L. quadribarbis, L. sanagaensis, L. sorex, and L. sp. nov. CO8-53, with pure CAs (nucleotides that are private to that species). Labeo lividus, L. nasus, and L. weeksii with compound CAs (combinations of nucleotides).
Discussion
It is noteworthy that 160 LCR species are represented by type specimens (wholly or in part) collected in, or in close vicinity of, the LCR (). The region is valuable for DNA barcoding then, not only for its high levels of endemism but also for its concentration of type localities for the 37 nonendemics described from the region. Verified barcodes of species collected in close proximity to the type locality (as discussed in the following section) are particularly valuable, as their legitimacy is less likely to be confounded in species where the range and species limits are obscure. From this perspective, a region that is unremarkable in terms of diversity could still be deserving of concerted efforts aimed at amassing barcodes due to its value as a locus of historically important collecting activity and taxonomic descriptions. Determining whether the species of uncertain taxonomic classification (denoted in with ‘?’) collected in LCR are novel localized endemics or isolated populations of extralimital species will require significant revisionary effort, including access to sequences from type or, more commonly, topotypical material (see discussion below for selected Labeo species).
After years of civil unrest and conflict (Prunier Citation2009; Reyntjens Citation2009), transportation infrastructure and road access to the LCR has declined markedly, particularly on the RC side of the main channel; many type localities (if known with any precision) are therefore no longer accessible. As a result of this inaccessibility, among other factors, much of the LCR remains unexplored ichthyologically. It is noteworthy that, despite intensive collecting efforts, 38 of the species (including 20 putative endemics) historically recorded as present in the LCR have yet to be recollected. Many of these species were not collected since they were originally described, and are known only from type specimens (e.g. Amphilius lamani Lönnberg and Rendahl 1920, Labeobarbus dartevellei Poll 1945, or Tylochromis praecox Stiassny 1989). While it may be the case that these species are indeed rare, it is more likely that they are highly localized endemics and/or habitat specialists, and as such have eluded capture due to vague locality records and the logistical difficulties of comprehensively surveying this dynamic system. The recent progress that was made here, as well as in select sites outside the LCR, suggest that the recorded diversity of fishes from within the Congo Basin can ultimately be expected to significantly exceed 1000 species.
Labeo case study
The addition of COI sequence data confirmed morphology-based species identifications for a majority of the taxa sampled in our study, but flags a number of others as being in need of further study. The pitfalls of assessing species relationships based on a COI (or any other single gene) tree are evident here, illustrated by the anomalous placement of L. batesii and L. annectens in the COI gene tree and the lack of strong bootstrap support at many nodes. When trees were built using rag1 + control region, as well as separate rag1 and control region trees, all agreed with the multi-gene topology, suggesting that the aberrant COI result was not a case of introgression. The utility of COI barcodes for identifying taxa in need of further taxonomic effort (e.g. collection, examination, or circumscription) is also evident. For Labeo, the COI tree topology was for the most part concordant with the three-gene tree.
In this respect, perhaps the greatest utility that we found working on this morphologically challenging group was in using DNA barcoding to highlight potential misidentifications. Throughout the course of the present study, problematic individuals were re-identified up to four times. Prior to the participation of the authority on African Labeo (SMT), a preliminary analysis of the samples revealed many apparently misplaced individuals. Having these specimens “flagged” optimized the limited time SMT could devote to identifying and describing hundreds of individuals, and demonstrates that barcoding can help to accelerate the pace of taxonomic research.
Morphological and DNA identifications for several putative species were in disagreement and require additional sampling and study. This is true as well for species represented by single vouchers, for which character-based COI may be polymorphic. Despite some differences in dorsal fin shape, scale counts, and lateral line coloration used to morphologically differentiate L. chariensis, L. parvus, L. cf. parvus, and L. simpsoni, our genetic results suggest that these characters may exhibit considerable variability, and as such may not provide an adequate basis for species delimitation. A putative novel species L. “longfin”, collected in LCR, for instance, shared the same haplotype with 10 individuals from LCR identified as L. simpsoni, and as such is probably best considered as a morphologically aberrant member of that species.
Examination of Labeo species is particularly instructive in demonstrating the potential for the FISH-BOL database to become corrupted by accessions sampled far from their species’ type locality. L. chariensis epitomizes this danger, as the type of this species is from the Shari River in the Chad Basin, some 1400 km north of the sampled regions. It is not improbable, therefore, that the L. chariensis assigned in this study (Lulua River), and the L. cf. parvus individuals (Lulua River), many of which share identical haplotypes, represent an altogether different species for which useful morphological diagnostics remain to be discovered. Similar reservations pertain to the divergent LCR and Lulua River specimens that, based on morphology, are all identified as L. simpsoni, but for which the type locality (the Chambezi River) is located on the other side of the Congo basin from the collection sites sampled for the present study. Determining which of these (if any) correspond with the Chambezi L. simpsoni must await further investigation, central to which will be molecular sampling of topotypical Chambezi L. simpsoni.
Recourse to molecular sampling of topotypical materials is needed for other taxa also. For example, the identity of the two specimens identified as L. cf. quadribarbis and collected from a single tributary of the LCR (the Mpozo River) needs corroboration. The types of L. quadribarbis are from the Yangambi region of the upper Congo (near Kisangani) and only two individuals of this species (MRAC 78078–78079) from Dilolo, Katanga, have been previously collected outside of this region. Again, reference to tissue vouchers from the vicinity of the type locality would be necessary to determine whether the LCR specimens represent a major range extension for this species, or whether they represent a morphologically similar, yet localized LCR endemic.
Our study also served to clarify some contrasting species divisions. Based on the concordance of distinct morphological and molecular characters, we herein recognize the validity of L. lividus and consider it to be another LCR endemic. We note, however, that this conclusion deserves additional scrutiny. For L. maleboensis and L. rectipinnis, conversely, the lack of unique molecular characters identified for those taxa suggests that further investigation could be helpful. The sole specimen of L. maleboensis shares an identical haplotype with L. lineatus. This is noteworthy as Tshibwabwa (Citation1997) lists a series of seemingly stable morphometric and meristic features that readily differentiate L. maleboensis from L. lineatus. Conversely, the validity of species currently recognized as synonyms for wide-ranging species such as L. coubie, L. lukulae, L. chariensis, and L. parvus need re-examination, as the divisions are probably overly broad for wide-ranging species and each probably represents a complex of morphologically cryptic species in need of further taxonomic resolution. Ward et al. (Citation2009) noted that more than five barcodes may need to be collected for freshwater species due to their greater spatial genetic heterogeneity (Ward et al. Citation1994), and this assertion is underscored by the lack of clarity regarding species limits with several taxa, once we include specimens sampled from outside the LCR. Moreover, for species where the current taxonomy is a “work in progress”, it will be critical for the purposes of establishing an authoritative FISH-BOL library that barcodes for described species be taken from as close to type localities as possible; otherwise, in many instances they are likely to create more confusion than they alleviate. Indeed, we would suggest that any topotypical sequences deposited in the BOLD library be annotated as such (as demonstrated in the present study), as these are of particular taxonomic importance.
Clearly, questions of Labeo species limits would benefit from more intensive geographic sampling and more detailed morphological investigation including osteological features (which may prove less intraspecifically variable than the standard meristic and fin form features that have traditionally been used in species delimitation), as well as computational image analysis and feature extraction (La Salle et al. Citation2009). Other corroborative evidence on these putative species’ natural history may be particularly helpful in better resolving Labeo taxonomy. Paugy et al. (Citation1990) found monogenean host fidelity in West African L. coubie and L. parvus, which they suggest can be employed as a third line of evidence along with genetic and morphometric data to validate species limits. Comparative parasitology (e.g. Locke et al. Citation2010) of Labeo would be an excellent application and case study for DNA barcoding.
Ultimate species resolution of fishes in the Lower Congo Basin will only be achieved by addressing the taxonomic impediment in Africa (Stiassny Citation2002). Taxonomy in the Congo region has withered due to half a century of unremitting political instability following independence (Reader Citation1999; Clark Citation2002). Expertise, museum collections, libraries, and the preponderance of opportunities in comparative biology remain centered in Europe or the USA—not in Central Africa, where these resources are most needed (Klopper et al. Citation2002; Agosti Citation2006; Swartz et al. Citation2008).
Digital repositories of taxonomic names, images, literature, and species attributes may help compensate for deficient research facilities in many parts of Africa and elsewhere (Miller Citation2007). Contributing specimen provenance and barcode sequence data to an online collaborative workbench for taxonomic research (e.g. BOLD) conceptually unites a diversity of collections and specimens under a common registry of genetic sequence accessions (Hanner and Gregory Citation2007) facilitating the use of these resources. The emergence of increasingly sophisticated cyber-infrastructure—coupled with more pluralistic approaches to integrative taxonomy (Padial et al. Citation2010)—augurs that a more accessible taxonomy is indeed on the horizon. Large-scale collaboration and data-sharing projects like this one are helping to realize this potential and can be an important element in the future development of more in situ taxonomic expertise.
Acknowledgements
We are extremely grateful to Sinaseli M. Tshibwabwa for his identification of the Labeo specimens used in this study. His efforts are greatly appreciated, and while he is in disagreement with some of the conclusions presented here, without his help we could not have carried out this study. The authors also thank H. Escobar, V. Mamonekene, J.J. Mbimbi, R. Monsembula, R. Schelly, and J. Schwarzer for field assistance, U. Schliewen (ZSM) for generously providing tissues, and WWF-Kinshasa for logistical support in the field. This manuscript benefited immensely from comments by S.O. Kolokotronis and an anonymous reviewer.
Declarations of interest: Funding for field research, collecting, and student training was provided by the National Science Foundation, through Biotic Surveys and Inventories award to M. L. J. S. (DEB Grant No. 0542540); the Axelrod Curatorship at AMNH (conferred on M. L. J. S.) provided additional funding to support S. M. T. on three study visits to examine Labeo specimens at the AMNH Ichthyology Collections. J. H. L. received funding from Columbia University's Department of Ecology, Evolution, and Environmental Biology, as well as a NSF Graduate Research Fellowship, in addition to support by the AMNH Axelrod Curatorship (conferred on M. L. J. S.). The authors acknowledge the International Barcode of Life Project (funded by Genome Canada via the Ontario Genomics Institute) and the Canadian Barcode of Life Network funded by the Natural Sciences and Engineering Research Council of Canada for informatics and sequencing support.
References
- Agosti D. 2006. Biodiversity data are out of local taxonomists' reach. Nature. 439:392.
- Amato G, Egan MG, Rabinowitz A. 1999. A new species of muntjac, Muntiacus putaoensis (Artiodactyla: Cervidae) from northern Myanmar. Anim Conserv. 2:1–7.
- Boulenger GA. 1901. Les poissons du bassin du Congo. Bruxelles: État Indépendant du Congo p 532.
- Boulenger GA. 1909. Catalogue of the fresh-water fishes of Africa in the British Museum of Natural History. London: Taylor and Francis p 373.
- Chapman L, Chapman C. 2001. Fishes of the African rain forests. In: Weber W, White LJT, Vedder A, Naughton-Treves L. editors. African rain forest ecology and conservation: An interdisciplinary perspective. New Haven, CT: Yale University Press263–290.
- Clark JF. 2002. The African Stakes of the Congo War. New York: Pellgrave MacMillan p 266.
- Davis JI, Nixon KC. 1992. Populations, genetic variation, and the delimitation of phylogenetic species. Syst Biol. 41:421–435.
- DeSalle R. 2006. What's in a character?. J Biomed Inform. 39:6–17.
- DeSalle R, Egan M, Siddall M. 2005. The unholy trinity: Taxonomy, species delimitation and DNA barcoding. Philos Trans R Soc London Ser B Biol Sci. 360:1905–1916.
- Gilles A, Lecointre G, Miquelis A, Loerstcher M, Chappaz R, Brun G. 2001. Partial combination applied to phylogeny of European cyprinids using the mitochondrial control region. Mol Phylogenet Evol. 19:22–33.
- Gordon D. 2006. Nachituti's gift: Economy, society, and environment in Central Africa. Madison, WI: University of Wisconsin Press.
- Hanner R, Gregory T. 2007. Genomic Diversity Research and the Role of Biorepositories. Cell Preservation Technology. 5:10.
- Hubert N, Hanner R, Holm E, Mandrak N, Taylor E, Burridge M, Watkinson D, Dumont P, Curry A, Bentzen P. 2008. Identifying Canadian freshwater fishes through DNA barcodes. PLoS ONE. 3:e2490 Doi: 2410.1371/journal.pone.0002490.
- Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. 2007. Universal primer cocktails for fish DNA barcoding. Molecular Ecology Notes. 7:544–548.
- Janzen D, Hallwachs W, Blandin P, Burns J, Cadiou J, Chacon I, Dapkey T, Deans A, Epstein M, Espinoza B. 2009. Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity. Molecular Ecology Resources. 9:1–26.
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9:286–298.
- Klopper R, Smith G, Chikuni A. 2002. The global taxonomy initiative in Africa. Taxon. 51:159–165.
- Lanave C, Preparata G, Saccone C, Serio G. 1984. A new method for calculating evolutionary substitution rates. J Mol Evol. 20:86–93.
- La Salle J, Wheeler Q, Jackway P, Winterton S, Hobern D, Lovell D. 2009. Accelerating taxonomic discovery through automated character extraction. Zootaxa. 2217:43–55.
- Lévêque C. 1997. Biodiversity dynamics and conservation: The freshwater fish of Tropical Africa. Cambridge: Cambridge University Press p 452.
- Locke S, Mclaughlin J, Marcogliese D. 2010. DNA barcodes show cryptic diversity and a potential physiological basis for host specificity among Diplostomoidea (Platyhelminthes: Digenea) parasitizing freshwater fishes in the St. Lawrence River, Canada. Mol Ecol. 19:2813–2827.
- López J, Chen W, Ortí G. 2004. Esociform phylogeny. Copeia. 2004:449–469.
- Lowenstein J, Amato G, Kolokotronis S. 2009. The real maccoyii: Identifying tuna sushi with DNA barcodes—contrasting characteristic attributes and genetic distances. PLoS ONE. 4:e7866.
- Markert J, Schelly R, Stiassny M. 2010. Genetic isolation and morphological divergence mediated by high-energy rapids in two cichlid genera from the lower Congo rapids. BMC Evol Biol. 10:149.
- Miller S. 2007. DNA barcoding and the renaissance of taxonomy. P Natl Acad Sci USA. 104:4775–4776.
- Miller M, Holder M, Vos R, Midford P, Liebowitz T, Chan L, Hoover P, Warnow T. 2009. The CIPRES Portal [Online]. Available at http://www.phylo.org [Accessed 3 June 2010].
- Nickum JG. 2004. Guidelines for the use of fishes in research. Bethesda, MD: American Fisheries Society.
- PAFFA. 2008. Workshop report In The First African Fish Barcode of Life initiative (FISH-BOL) Regional Working Group Workshop. Addis Ababa: Addis Ababa University, Ethiopia.
- Padial J, Miralles A, De la Riva I, Vences M. 2010. The integrative future of taxonomy. Front Zool. 7:16.
- Paugy D, Guégan J, Agnèse J. 1990. Three simultaneous and independent approaches to the characterization of a new species of Labeo (Teleostei, Cyprinidae) from West Africa. Can J Zool. 68:1124–1131.
- Prunier G. 2009. Africa's World War: Congo, the Rwandan Genocide, and the Making of a Continental Catastrophe. Oxford: Oxford University Press.
- Rambaut A. 2010. FigTree v1.3.1 [Online]. Available at http://tree.bio.ed.ac.uk [Accessed 30 October 2010].
- Ratnasingham S, Hebert PDN. 2007. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol Ecol Notes. 7:355–364.
- Reader J. 1999. Africa: A biography of the continent. New York: Vintage p 816.
- Reid GM. 1985. A revision of African species of Labeo (Pisces: Cyprinidae) and a re-definition of the genus. Braunschweig: Verlag van J. Crammer p 322.
- Reyntjens F. 2009. The Great African War: Congo and Regional Geopolitics, 1996–2006. Cambridge: Cambridge University Press p 340.
- Roberts T. Review: A revision of African species of Labeo (Pisces: Cyprinidae) and a re-definition of the genus. G.M. Reid 1985. Copeia. 1986847–848.
- Roberts T, Stewart D. 1976. An ecological and systematic survey of fishes in the rapids of the Lower Zaire or Congo River. Bull Mus Comp Zool Harvard Coll. 147:239–317.
- Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 132:365–386.
- Runge J. 2007. The Congo River, Central Africa. In: Gupta A. editors. Large rivers: Geomorphology and management. New York: Wiley293–309.
- Sarkar IN, Thornton JW, Planet PJ, Figurski DH, Schierwater B, DeSalle R. 2002. An automated phylogenetic key for classifying homeoboxes. Mol Phylogenet Evol. 24:388–399.
- Skelton P. 1994. Diversity and distribution of freshwater fishes in East and Southern Africa. In: Teugels GG, Guégan JF, Albaret JJ. editors. Biological diversity of African fresh and brackish water fishes. Tervuren: Musee Royal de l'Afrique Centrale95–131.
- Skelton P, Tweddle D, Jackson P. 1991. Cyprinids of Africa. In: Winfield I, Nelson J. editors. Cyprinid fishes, systematics, biology and exploitation. London: Chapman and Hall211–233.
- Stamatakis A. 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57:758–771.
- Stiassny MLJ. 2002. Conservation of freshwater fish biodiversity: The knowledge impediment. Verh Gesellsch Ichthyol. 3:7–18.
- Stiassny MLJ, Getahun A. 2007. An overview of Labeonin relationships and the phylogenetic placement of the Afro-Asian genus Garra Hamilton, 1922 (Teleostei: Cyprinidae), with the description of five new species of Garra from Ethiopia, and a key to all African species. Zool J Linnean Soc. 150:41–83.
- Swartz ER, Mwale M, Hanner R. 2008. A role for barcoding in the study of African fish diversity and conservation. S Afr J Sci. 104:293–298.
- Thieme ML, Abell R, Stiassny MLJ, Skelton PH. 2005. Freshwater ecoregions of Africa and Madagascar: A conservation assessment. Washington, DC: Island Press p 431.
- Thys van den Audenaerde D. 1987. Review of Reid GM 1985, a revision of African species of Labeo (Pisces, Cyprinidae). Rev Zool Afr. 100:487–492.
- Tshibwabwa SM. 1997. Systématique des espéces africaines du genre Labeo (Teleostei, Cyprinidae) dans les régions ichtyogéographiques de Basse-Guinée et du Congo Vol I et II. Faculté des Sciences, Facultés Universitaires Notre-Dame de la Paix de Namur, Belgium. Namur: Presses Universitaires de Namur p 530.
- Tshibwabwa SM, Teugels G. 1995. Contribution to the systematic revision of the African cyprinid fish genus Labeo: Species from the Lower Zaire river system. J Nat Hist. 29:1543–1579.
- Tshibwabwa SM, Stiassny MLJ, Schelly R. 2006. Description of a new species of Labeo (Teleostei: Cyprinidae) from the Lower Congo River. Zootaxa. 1224:33–44.
- Victor B. 2007. Coryphopterus kuna, a new goby (Perciformes: Gobiidae: Gobiinae) from the western Caribbean, with the identification of the late larval stage and an estimate of the pelagic larval duration. Zootaxa. 1526:51–61.
- Ward RD, Woodwark M, Skibinski DOF. 1994. A comparison of genetic diversity levels in marine, freshwater, and anadromous fishes. J Fish Biol. 44:213–232.
- Ward RD, Hanner R, Hebert P. 2009. The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol. 74:329–356.
- Wong E, Shivji H, Hanner R. 2009. Identifying sharks with DNA barcodes: Assessing the utility of a nucleotide diagnostic approach. Mol Ecol Resources. 9:243–256.
- Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol. 39:306–314.
