Abstract
Background and aims. We analyzed a small and wide geographically distributed Neotropical freshwater fish, the Piabina argentea from the Upper Paraná Basin, to check the hypothesis that this species is composed of more than one biological unit, since it has a limited dispersion, through the DNA barcode technique. Materials and methods. Partial mitochondrial COI and CytB gene sequences were obtained for 58 specimens drawn from 13 localities. Results. Phylogenetic analysis revealed six major clusters of P. argentea. Kimura-two-parameter (K2P) genetic divergences among these six P. argentea clusters ranged from 2 to 5.6% and from 2.3 to 5.4% for COI and CytB genes, respectively, and these values were on average approximately nine times greater than intra-cluster K2P divergences. The fixation index (FST) among clusters showed very high values and the haplotype network analysis displayed seven unconnected units. Conclusion. These results reinforce the hypothesis that the widely distributed P. argentea species concept as currently conceived actually represents more than one species (possibly six). These results demonstrate the efficacy of DNA barcoding for the discovery of hidden diversity in Neotropical freshwater fishes, and we conclude that barcoding is a useful tool for alpha taxonomy.
Introduction
The Neotropical freshwater fish fauna is one of the richest in the world (Schaefer Citation1998), with about 6000 species recognized in this region, out of which 4475 are actually considered valid and 1550 are recognized but not yet described (Reis et al. Citation2003). In Brazil, there are about 2587 valid species and many others to be described (Buckup et al. Citation2007), but, even so, the sampling of species is insufficient and many regions remain almost unexplored (Langeani et al. Citation2006; Junk Citation2007). Schaefer (Citation1998) estimates that there may be as many as 8000 species in the Neotropical region. For example, in a recent study of the fish fauna from the Upper Paraná Basin, the best studied region in Neotropical area, Langeani et al. (Citation2006) made an inventory, which revealed that about 15% (∼50) represent new species. Many other works pointed that the number of fish species tends to increase mainly among those fish belonging to small-sized groups and that inhabit headwaters streams (Schaefer Citation1998; Vari and Malabarba Citation1998; Castro et al. Citation2003, Citation2004, Citation2005; Langeani et al. Citation2006). Additionally, the geographic distribution pattern of the Neotropical fish species is very complex, with some species having a very restricted distribution (e.g. Trichomycterus maracaya, Characidium xanthopterum) occurring mostly in headwaters, and others having a wide distribution (e.g. Hoplias malabaricus, Astyanax paranae) sometimes occurring in more than one hydrographic basin (Reis et al. Citation2003; Junk Citation2007).
The genus Piabina, composed by small fishes (∼50 mm), belongs to the family Characidae but its relationship with the remaining characids is uncertain (Lima et al. Citation2003). Two species are assigned to Piabina: Piabina argentea Reinhardt, 1867 and Piabina anhembi da Silva and Kaefer, Citation2003. P. argentea has a wide geographic distribution occurring in the Upper Paraná Basin (the same region of this work); in the São Francisco Basin (type-locality); and in the Itapirucu, Paraíba do Sul, and Itapemirim rivers (eastern Brazilian basins) (Vari and Harold Citation2001). P. anhembi is restricted to its type-locality (Upper Tietê River, Salesópolis, São Paulo, Brazil) (da Silva and Kaefer Citation2003). These two species differ from each other by the teeth position, head size, and mouth proportions (da Silva and Kaefer Citation2003). Piabina differs from its putative sister group, Creagrutus, only by two subtle characters: the fourth infraorbital bone morphology and the teeth position (Vari and Harold Citation2001). Creagrutus and Piabina were allopatric (Vari and Harold Citation2001) until the discovery of a new Creagrutus species in the Upper Paraná River Basin (Ribeiro et al. Citation2004). The Piabina species populations have a limited dispersion, usually living in a restricted hydrographic region (Lowe-Mcconnell Citation1999). Castro (Citation1999) suggests a limited dispersion to small fishes, which restricts their geographical distribution and may facilitate the population geographical subdivision enabling the possible creation of new species by geographic isolation (allopatry).
The advance of molecular techniques has proven a useful tool in biodiversity studies, mainly in those cases where the traditional tools are insufficient or unable to identify species. The use of genetic techniques has revealed that some species are actually species complexes (Agostinho et al. Citation2007). Bickford et al. (Citation2006) showed that there has been an increased recognition of cryptic species from different groups of animals and plants in the past two decades due to the use of molecular methods. Hebert et al. (Citation2003) proposed the DNA barcoding technique as a useful molecular tool for the identification of species, and many published works have shown the efficacy of this methodology for the identification of several organisms (Hebert et al. Citation2004a; Ward et al. Citation2005; Clare et al. Citation2007; Kelly et al. Citation2007; Hubert et al. Citation2008; Valdez-Moreno et al. Citation2009). Hebert et al. (Citation2004b) proposed a threshold to delimit species that are 10 × larger than the intraspecific average values. New species have been proposed with DNA barcoding data and some of these species have been formally described later (Smith et al. Citation2005; Witt et al. Citation2006; Ward Citation2007; Nguyen and Seifert Citation2008; Ward et al. Citation2008; Yassin et al. Citation2008).
Considering the wide distribution and limited dispersion of small fish P. argentea and the promising use of DNA barcodes for flagging new species, the present work assessed samples of P. argentea from the Upper Paraná and São Francisco basins to check the hypotheses that this species could represent more than one biological unit.
Materials and methods
Specimen collection
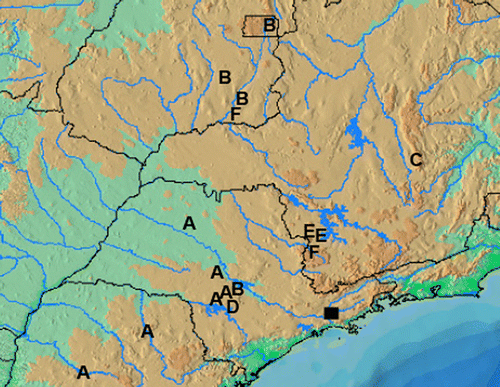
Fifty-three P. argentea specimens from 12 sites located in the Upper Paraná Basin and one in the São Francisco Basin and five P. anhembi specimens from the Upper Paraná River Basin were collected ( and ). The Velhas River in the São Francisco Basin was sampled because this is the type locality of P. argentea. Additionally, two Creagrutus specimens (Creagrutus meridionalis and Creagrutus paraguayensis) from the Paraguay River Basin were used as outgroup (). All specimens had a fresh fragment tissue removed and preserved in absolute ethanol at − 20°C. Voucher specimens were deposited in the collection of Laboratório de Biologia e Genética de Peixes, Departamento de Morfologia, Instituto de Biociências, UNESP, Botucatu, São Paulo, Brazil. The specimens' provenance data were deposited in BOLD Project EFUPR (Ratnasingham and Hebert Citation2007).
Table I. Specimen data.
Extraction, PCR, and sequencing
Total genomic DNA was isolated from fin or muscle tissue of each specimen using the DNeasy Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The partial mitochondrial cytochrome c oxidase subunit I gene (COI, 648 bp) was amplified by the PCR using two sets of primers: FishF1, 5′-TCAACCAACCACAAAGACATTGGCAC-3′; FishF2, 5′-TCGACTAATCATAAAGATATCGGCAC-3′; FishR1, 5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′; and FishR2, 5′-ACTTCAGGGTGACCGAAGAATCAGAA-3′ (Ward et al. Citation2005). The whole cytochrome b (CytB, 1118 bp) mitochondrial gene was amplified by PCR using the CytB-F, 5′-GACTTGAAAAACCAYCGTTGT-3′, and CytB-R, 5′-GCTTTGGGAGTTAGDGGTGGGAGTTAGAATC-3′ (C. Oliveira, pers. comm.). PCR was carried out on a thermocycler (Veriti® 96-Well Thermal Cycler; Applied Biosystems, Foster City, California, USA) with a final volume of 12.5 μl containing 0.3 μl dNTP (2 mM), 1.25 μl 10 × Taq buffer (50 mM KCl, 10 mM Tris–HCl, 0.1% Triton X-100, and 1.5 mM MgCl2), 0.3 μl each primer (10 μM), 0.7 μl MgCl2 (50 mM), 0.05 μl Taq-Pht DNA polymerase (5 U), 1 μl template DNA (10–20 ng), and ultrapure water. The thermocycler conditions to amplify the COI were initial denaturation at 95°C for 5 min followed by 30 cycles of denaturation at 95°C for 45 s, annealing at 55°C for 30 s, and extension at 72°C for 60 s. A final extension at 72°C for 10 min was performed. The thermocycler conditions to amplify the CytB were initial denaturation at 95°C for 5 min followed by two cycles of denaturation at 95°C for 30 s, annealing at 55°C for 45 s, and extension at 72°C for 60 s; two cycles of denaturation at 95°C for 30 s, annealing at 50°C for 45 s, and extension at 72°C for 60 s; two cycles of denaturation at 95°C for 30 s, annealing at 48°C for 45 s, and extension at 72°C for 60 s; 25 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 45 s, and extension at 72°C for 60 s; and a final extension at 72°C for 5 min. Amplified products were checked on 1% agarose gels stained with Blue Green Loading Dye I (LGC Biotecnologia, Cotia, Sa˜o Paulo, Brazil). The PCR products were purified with ExoSAP-IT® (USB Corporation, Cleveland, OH, USA) following the manufacturer's protocol. The purified PCR product was used as template to sequence both DNA strands. The cycle sequencing reaction was carried out using a BigDye™ Terminator v3.1 Cycle Sequencing Ready Reaction kit (Applied Biosystems) in a final volume of 7 μl containing 1.4 μl template, 0.35 μl primer (10 μM), 1.05 μl buffer 5 × , 0.7 μl BigDye mix, and water. The cycle sequencing conditions were initial denaturation at 96°C for 2 min followed by 30 cycles of denaturation at 96°C for 45 s, annealing at 50°C for 60 s, and extension at 60°C for 4 min. The PCR sequencing products were purified with ethylenediamine tetraacetic acid/sodium acetate/alcohol following the protocol suggested in the BigDye™ Terminator v3.1 Cycle Sequencing kit's manual (Applied Biosystems). All samples were sequenced on an ABI 3130 Genetic Analyzer (Applied Biosystems) following the manufacturer's instructions. All sequences were deposited in the GenBank and in the Barcode of Life Data Systems (Project EFUPR) ().
Data analysis
All sequences were analyzed using SeqScape® software v2.6 (Applied Biosystems) to obtain the consensus sequences and check the occurrence of deletions, insertions, and stop codons. The sequences were aligned using the online version of MUSCLE (Edgar Citation2004). The genetic distance among and within observed clusters was calculated using the Kimura-two-parameter (K2P) distance model (Kimura Citation1980) for both genes separately. A neighbor-joining (NJ) tree of K2P distances using the combined COI and CytB sequences was created to provide a graphic representation of the relationships among specimens and clusters with the software MEGA 4.0 (Tamura et al. Citation2007). Bootstrap resampling (Felsenstein Citation1985) was applied to assess the support for individual nodes using 1000 pseudo-replicates.
Phylogenetic analyses using maximum parsimony were performed using PAUP* version 4.0b10 (Swofford Citation2002) with heuristic searches, random addition of sequences, and tree bisection and reconnection algorithms. The ACCTRAN optimization method was utilized. The parsimony trees were constructed using a 1:1 transition–transversion ratio. Cluster robustness was assessed using 1000 bootstrap pseudo-replicates (Felsenstein Citation1985) with the same parameters cited above.
The seven major clusters obtained were considered as different units for the fixation index (FST) calculation using Arlequin 3.11 (Excoffier et al. Citation2005). A statistical parsimony network was constructed using TCS 1.21 (Clement et al. Citation2000), which employs the method of Templeton et al. (Citation1992) with a statistical confidence interval of 90%. The analyses were carried out in TCS using the “fix connection limit” option to obtain the mutational steps necessary to connect the seven observed haplotype networks. The ancestral haplotype was also identified using TCS according to the method of Castelloe and Templeton (Citation1994).
Results
Sequence data
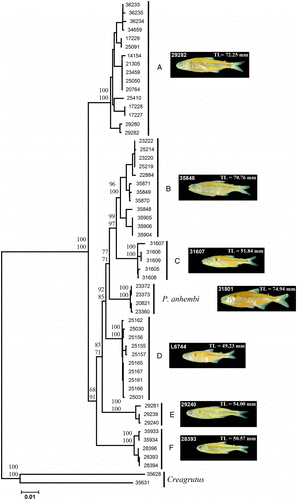
Sequence data for a 648 bp fragment of COI and 1118 bp of CytB were obtained for a total of 58 Piabina specimens (53 P. argentea and 5 P. anhembi). We also obtained the COI and CytB sequences from two specimens of Creagrutus (C. meridionalis and C. paraguayensis) used as an outgroup. No sequences showed insertions, deletions, or stop-codons, and the global transition–transversion ratio was 4.4. A total of 233 nucleotides (72 in COI and 161 in CytB) were variable in the data set of Piabina specimens (∼13%—outgroup not considered) and 209 of them were informative in the parsimony analyses. These variations defined a total of 42 haplotypes (COI and CytB displayed 28 and 39 haplotypes, respectively). The two methods of tree construction (NJ and maximum parsimony) resulted in the same topology (except for some internal taxa in the subclusters; data not shown), which showed seven major clusters with high support values (). P. anhembi samples formed one cluster and P. argentea samples were divided into six clusters, one corresponding to the sample from the São Francisco River Basin (Cluster C) and five clusters representing P. argentea samples from the Upper Paraná Basin (). These seven clusters are divided into two major groups, one containing Cluster A and a second group with the other clusters (). We use two different methodologies of tree construction to check the robustness of the data.
Figure 2. NJ tree of COI/CytB showing the seven major clusters obtained among Piabina specimens (A–F represent P. argentea). Node values represent statistic support: upper values, NJ bootstrap (1000 pseudo-replicates); lower values, maximum parsimony bootstrap (1000 pseudo-replicates). Numbers on fishes represent voucher number and size of photographed specimens (left and right, respectively).
The inter-cluster K2P genetic distance values ranged from 2% (Clusters D × E) to 5.6% (Clusters A × C) and from 2.3% (Clusters B × C) to 5.4% (Clusters A × E) for COI and CytB, respectively (). The average intra-cluster K2P distance ranged from 0 to 0.9% (average = 0.36%) for COI and from 0.1 to 1% (average = 0.5%) for CytB ().
Table II. K2P genetic distance obtained among the seven major Piabina clusters.
Cluster comparisons
The pairwise FST index among the seven clusters identified showed values from 0.77 to 0.98 for COI and from 0.66 to 0.96 for CytB, all highly significant (p < 0.001) ().
Table III. Pairwise FST index obtained among the seven major Piabina clusters.
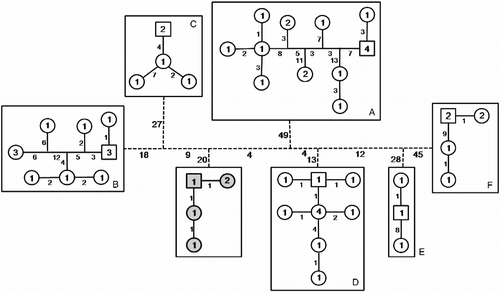
The haplotype network based on Templeton's method (Templeton et al. Citation1992) with the combined data set (COI/CytB) displayed seven unconnected networks, one representing P. anhembi and the other six representing P. argentea (). This result is consistent with the seven clusters identified through the phylogenetic analysis (). The number of haplotypes present in each network range from 3 (Cluster E) to 11 (Cluster A) (), and the number of mutational steps necessary to connect the independent P. argentea networks ranged from 45 to 110 (, dashed lines). The haplotype network was constructed for each separate gene to check whether the same seven unconnected networks would be obtained. Both genes displayed the same result, with 14-34 (COI) and 25-71 (CytB) mutational steps necessary to connect the independent networks (networks not shown).
Figure 3. Seven unconnected haplotype networks among Piabina specimens. P. anhembi is represented in gray. Numbers inside the figures represent specimens that share the same haplotype. Numbers on lines represent the mutational steps between haplotypes. Dashed lines represent the necessary steps to connect the independent networks.
Discussion
The specimens of Piabina were divided into seven clusters in the phylogenetic analysis, one cluster representing P. anhembi and the six other representing P. argentea (). The data showed the absence of genetic flow among local samples and permit one to suggest that P. argentea represents six different biological units (meaning a minimum of five new species). These seven clusters were confirmed by haplotype network () and are divided into two major groups (). The first group contains the Cluster A and is the sister group of the second group, composed by the remaining clusters, including P. anhembi (). The average inter-cluster K2P distance values among P. argentea were about nine times greater than the average intra-cluster values found for the COI (from 5.6 to 15.6 × ) and CytB genes (from 4.6 to 10.8 × ) () and the inter-cluster values among the P. argentea units were similar to the values between P. argentea clusters and their congeners P. anhembi (average = 3.0 and 3.8% for COI and CytB, respectively), reinforcing the hypothesis of the existence of more than one biological unit for P. argentea (). These results corroborate the hypothesis of limited dispersion for Piabina species (Lowe-Mcconnell Citation1999) and other small fishes (Castro Citation1999), which facilitates the population geographical subdivision enabling the possible creation of new species by geographic isolation (allopatry).
Hebert et al. (Citation2004b) suggested a threshold to delimit species with DNA barcode data. These values should be at least 10 × the average intraspecific values. The average intra-cluster values of the six P. argentea clusters were 0.4% and 0.56% for COI and CytB, respectively, and some inter-cluster divergences within P. argentea are slightly below this limit (see ). However, a recent review of “barcoded” fishes (Ward Citation2009) noted that about 17% of the genetic divergence values among congeneric species were less than 3% divergent and that a further 3.7% of congeners are less than 1% divergent. The author suggests that if the unknown specimen is more than 2% divergent from the known specimen, it is very likely that this is a different species with a probability greater than 95%. Hence, the threshold limit proposed by Hebert et al. (Citation2004b) as an indicator of cryptic speciation should be carefully analyzed for each group.
Ward et al. (Citation2007), working with sharks of the genus Squalus, observed the formation of two clusters in the species Squalus acanthias, which showed a genetic divergence of just 0.76% between them. Interestingly, these two groups had been considered as two species until the decade of 1960: S. acanthias from the Atlantic and South Pacific Oceans and Squalus suckkeyi from the North Pacific Ocean (see Jensen Citation1966). The authors suggested the revalidation of the second species. The comparison with values among congener species may be useful for the delimitation of a threshold. Ornelas-Garcia et al. (Citation2008), working with species of the genus Astyanax from Mesoamerica, found that some specimens formed separate clusters and suggested the occurrence of a species complex in this genus, assigning provisional names to each cluster obtained. Ward et al. (Citation2008), working with Asian sea bass Lates calcarifer specimens from different localities (Australia and Myanmar), found genetic distance values of 9.5% between two groups for COI (DNA barcode region) and 11.3% for CytB. The authors suggested the existence of two species. The average divergence value of “barcoded” congeneric fishes is about 8.4% (Ward Citation2009). Values smaller than this average, such as those observed in the present work and in the above-cited papers, can be explained in two ways: the rate of evolution can vary among different higher taxa and, consequently, the accumulation of substitutions can vary. In fact, it has been observed that different teleost orders have different evolutionary rates (Krieger and Fuerst Citation2002). Another possible explanation could relate to species ages, where evolutionarily “young” species may not have had sufficient time to accumulate many mutations in their barcodes. In fact, Montoya-Burgos (Citation2003), working with species of Hypostomus from South America, suggested that the process of divergence and radiation in this genus dates back to sometime between 12 and 4 million years ago. Hubert et al. (Citation2007), working with Serrasalmus and Pygocentrus from South America, encountered similar values suggesting that species separation dates back to sometime between 8 and 2 million years ago. Both authors suggested that this pattern is valid for most Neotropical freshwater fishes. In their studies of Rhamdia and Synbranchus fish species, Perdices et al. (Citation2002, Citation2005) proposed similar patterns for Mesoamerica and Ornelas-Gacia et al. (Citation2008) corroborated the same patterns for Astyanax. With increasing recognition that mitochondrial DNA is under strong selection, some authors caution against the use of mitochondrial DNA data for dating divergence events, but, this caveat notwithstanding, recognize that selective sweeps can be beneficial for barcoding (Galtier et al. 2009). Molecular clock approaches that infer age of the most recent common ancestor tend to be overestimated using mitochondrial DNA unless they correct for apparent rate differences between short and long time frames (Rand 2008).
The intercluster analysis performed confirmed the presence of seven dissimilar barcode sequence clusters among the Piabina specimens examined. The haplotype networks obtained using the combined data set () and those for each gene separately (data not shown) displayed seven unconnected networks with high numbers of mutational steps (ranged from 45 to 110; ) necessary to connect these independent networks. This situation is not expected when the specimens represent a single species (Hart and Sunday Citation2007), even when there is very strong structure among populations. Some pairs of P. argentea clusters need more mutational steps than others to connect with their congener P. anhembi species (). Thus, these results support the hypothesis that P. argentea comprises more than one biological species. Kon et al. (Citation2007), working with the gobioid fish Schindleria, obtained an unconnected haplotype network with seven independent clusters and suggested that Schindleria represents a species complex, as imparted here.
The FST index showed very high values among the seven clusters obtained (), with similar values among P. argentea and P. anhembi clusters. Considering that FST values between 0 and 0.05 indicate a low genetic structure, values between 0.05 and 0.15 a moderate genetic structure, values between 0.15 and 0.25 a high genetic structure, values above 0.25 a strong genetic structure, and values close to 1 are usually found among different species (Wright Citation1978; Hartl and Clark Citation1997); the values presented in strongly suggest that our seven clusters represent different species.
Many species have been discovered with the use of molecular data and some have been formally described later (Smith et al. Citation2005; Witt et al. Citation2006; Kon et al. Citation2007; Ward et al. Citation2007, Citation2008; Nguyen and Seifert Citation2008; Yassin et al. Citation2008), and the DNA barcode has also been utilized as part of the validation and formal description of new fish species such as Coryphopterus kuna (Victor Citation2007); Urolophus kapalensis (Yearsley and Last Citation2006); Brachionichthys autralis (Last et al. Citation2007); five new species of Chromis genus (Pyle et al. Citation2008), Dipturus argentinensis (Diaz de Astarloa et al. Citation2008), and Moenkhausia forestii (Benine et al. Citation2009). Our data suggest that the widely distributed P. argentea species represent more than one biological unit in the Upper Paraná Basin, and probably this hypothesis is valid all over the area of occurrence of this species. Interestingly, some clusters were found only in a single locality (Clusters C–F, and P. anhembi) while others are widely dispersed (Clusters A and B) (). The fact that Clusters A and B are widely dispersed could be a cause of no prior recognition of these possible species, since the area of overlap between them could impede its recognition. Thus, we suggest that a detailed review of Piabina be conducted to validate these new species (sensu Padial et al. Citation2010). On the other hand, we believe that the analysis of many other widely distributed fish species may also disclose new species.
Conclusions
Our data demonstrate the efficacy of DNA barcoding for discriminating known species and to flag new ones, alone or associated with other genes. Despite the concerns of Hickerson et al. (Citation2006) to the contrary, DNA barcoding revealed the existence of separate taxa with low divergence rate or recent radiation. We also substantiate the use of DNA barcode sequences as part of the formal description of species. These data can be useful when morphological characters are insufficient or too weak to define a species and, importantly, because they apply to any sex or life stage, can help to disambiguate the application of names in future studies.
Acknowledgements
The authors are grateful to Renato Devidé and Ricardo Teixeira for their help with the fish collection. Financial support for the present study was provided by CNPq and FAPESP.
Declaration of interest: Financial support for this study was provided by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq).
References
- Agostinho AA, Pelicice FM, Petry AC, Gomes LC, Júlio HFJr. 2007. Fish diversity in the Upper Paraná River Basin: Habitats, fisheries, management and conservation. Aquat Ecosyst Health Manag. 10 2: 174–186.
- Benine RC, Mariguela TC, Oliveira C. 2009. Nem species of Moenkhausia Eigenmann, 1903 (Characiformes: Characidae) with comments on the Moenkhausiaoligolepis species complex. Neotrop Ichthyol. 7 2: 161–168.
- Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, . 2006. Cryptic species as a window on diversity and conservation. Trends Ecol Evol. 22 3: 148–155.
- Buckup PA, Menzes NA, Ghazzi MS. 2007. Catálogo das espécies de peixes de água doce do Brazil. Rio de Janeiro, Brazil: Museu Nacional.
- Castelloe J, Templeton AR. 1994. Root probabilities for intraspecific gene trees under neutral coalescent theory. Mol Phylogenet Evol. 3:102–113.
- Castro RMC. 1999. Evolução da ictiofauna de riachos sul-americanos: Padrões gerais e possíveis processos causais. In: Caramashi EP, Mazzoni R, Peres-Neto PR. editors. Ecologia de Peixes de Riacho: Série Oecologia Brasiliensis. Rio de Janeiro, Brazil: PPGE-UFRJ. 139–155.
- Castro RMC, Casatti L, Santos HF, Ferreira KM, Ribeiro AC, Benine RC, Dardis GZP, Melo ALA, Stopligia R, Abreu TX, Bockmann FA, Carvalho M, Gibran FZ, Lima FCT. 2003. Estrutura e composição da ictiofauna de riachos do Rio Paranapanema, sudeste e sul do BrazilBiota Neotropica. 3. Available at: http://www.biotaneotropica.org.br/v3n1/pt/abstract?article+BN0170301. Accessed on 15 July 2010.
- Castro RMC, Casatti L, Santos HF, Melo ALA, Martins LSF, Ferreira KM, Gibran FZ, Benine RC, Carvalho M, Ribeiro AC, Abreu TX, Bockmann FA, Pelição GZ, Stopligia R, Langeani F. 2004. Estrutura e composição da ictiofauna de riachos da bacia do Rio Grande, no Estado de São Paulo, Sudeste do Brazil. Biota Neotropica. 4 1 Available at: http://www.biotaneotropica.org.br/v4n1/pt/abstract?article+BN0170402004. Accessed on 15 July 2010.
- Castro RMC, Casatti L, Santos HF, Vari RP, Melo ALA, Martins LSF, . 2005. Structure and composition of the stream ichthyofauna of four tributary rivers of the Upper Rio Paraná Basin, Brazil. Ichthyol Explor Freshwaters. 16 3: 193–214.
- Clare EL, Lim BK, Engstrom MD, Eger JL, Hebert PDN. 2007. DNA barcoding of Neotropical bats: Species identification and discovery within Guyana. Mol Ecol Notes. 7:184–190.
- Clement M, Posada D, Crandall KA. 2000. TCS: A computerprogram to estimate gene genealogies. Mol Ecol. 9:1657–1659.
- da Silva JFP, Kaefer CC. 2003. Uma nova espécie de Piabina Reinhardt, 1867 (Teleostei: Ostariophysi: Characidae) para o alto Rio Tietê, São Paulo, Brazil. Comun Mus Ciênc Technol PUCRS Sér Zool Porto Alegre. 16 1: 53–65.
- Diaz de Astarloa JM, Mabragana E, Hanner R, Figueroa DE. 2008. Morphological and molecular evidence for a new species of longnose skate (Rajiformes: Rajidae: Dipturus) from Argentinean waters based on DNA barcoding. Zootaxa. 1921:35–46.
- Edgar RC. 2004. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 5:113.
- Excoffier L, Laval G, Schneider S. 2005. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinformatics Online. 1:47–50.
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 39:783–791.
- Galtier N, Nabholz S, Glémin S, Hurst GDD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology. 18:4541–4550.
- Hart MW, Sunday J. 2007. Things fall apart: Biological species form unconnected parsimony networks. Biol Lett. 3:509–512.
- Hartl DL, Clark AG. 1997. Principles of population genetics. Sunderland, MA: Sinauer Associates.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc Lond Ser B Biol Sci. 270:313–321.
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004a; 2:1657–1663.
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the Neotropical skipper butterfly Astrapesfulgerator. Proc Natl Acad Sci U S A. 2004b; 101 41: 14812–14817.
- Hickerson MJ, Meyer CP, Moritz C. 2006. DNA barcoding willoften fail to discover new animal species over broad parameter space. Syst Biol. 55:729–739.
- Hubert N, Duponchelle F, Nuñez J, Garcia-Davila C, Paugy D, Renno JF. 2007. Phylogeography of the piranha genera Serrasalmus and Pygocentrus: Implications for the diversification of the Neotropical ichthyofauna. Mol Ecol. 16:2115–2136.
- Hubert N, Hanner R, Holm E, Mandrak NE, Taylor E, Burridge M, . 2008. Identifying Canadian freshwater fishes through DNA barcodes. PLoS ONE. 3 6: e2490.
- Jensen AC. 1966. Life history of the spiny dogfish. Fish Bull. 65:527–554.
- Junk W. 2007. Freshwater fishes of South America: Their biodiversity, fisheries, and habitats: A synthesis. Aquat Ecosyst Health Manag. 10 2: 228–242.
- Kelly RP, Sarkar IN, Eernisse DJ, DeSalle R. 2007. DNA barcoding using chitons (genus Mopalia). Mol Ecol Notes. 7:177–183.
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Kon T, Yoshino T, Mukai T, Nishida M. 2007. DNA sequences identify numerous cryptic species of the vertebrate: A lesson from the gobioid fish Schindleria. Mol Phylogenet Evol. 44:53–62.
- Krieger J, Fuerst PA. 2002. Evidence for a slowed rate of molecular evolution in the order Acipenseriformes. Mol Biol Evol. 19:891–897.
- Langeani F, Castro RMC, Oyakawa OT, Shibatta OA, Pavanelli CS, Casatti L. 2006. Diversidade da fauna do Alto Rio Paraná: Composição atual e perspectivas futuras. Biota Neotrop. 7 3: 181–197.
- Last PR, Gledhill DC, Holmes BH. 2007. A new handfish, Brachionichthysaustralis sp. nov. (Lophiiformes: Brachionichthyidae), with a redescription of the critically endangered spotted handfish, B. hirsutus (Lacépède). Zootaxa. 1666:53–68.
- Lima FCT, Malabarba LR, Buckup PA, Pezzi da Silva JF, Vari RP, Harold A, . 2003. Genera Incertae Sedis in Characidae. In: Reis RE, Kullander SO, Ferraris CJJr. editors. Checklist of the freshwater fishes of South and Central America. Porto Alegre, Brazil: Edipucrs. 106–168.
- Lowe-Mcconnell RH. 1999. Estudos ecológicos de comunidades de peixes tropicais. São Paulo, Brazil: EDUSP.
- Montoya-Burgos JI. 2003. Historical biogeography of the catfish genus Hypostomus (Siluriformes: Loricariidae), with implications on the diversification of Neotropical ichthyofauna. Mol Ecol. 12:1855–1867.
- Nguyen HDT, Seifert KA. 2008. Description and DNA barcoding of three new species of Leohumicola from South Africa and the United States. Personia. 21:57–98.
- Ornelas-Garcia CP, Dominguez-Dominguez O, Doadrio I. 2008. Evolutionary history of the fish genus Astyanax Baird & Girard (1854) (Actynopterigii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evol Biol. 8:340.
- Padial JM, Miralles A, De la Riva I, Vences M. 2010. The integrative future of taxonomy. Front Zool. 7:16.
- Perdices A, Bermingham E, Montilla A, Doadrio I. 2002. Evolutionary history of the genus Rhamdia (Teleostei: Pimelodidae) in Central America. Mol Phylogenet Evol. 25:172–189.
- Perdices A, Doadrio I, Bermingham E. 2005. Evolutionary history of the synbranchid eels (Teleostei: Synbranchidae) in Central America and the Caribbean islands inferred from their molecular phylogeny. Mol Phylogenet Evol. 37:460–473.
- Pyle RL, Earle JL, Greene BD. 2008. Five new species of the damselfish genus Chromis (Perciformes: Labroidei: Pomacentridae) from deep coral reefs in the tropical western Pacific. Zootaxa. 1671:3–31.
- Rand DM. 2008. Mitigating mutational meltdown in mammalian mitochondria. PLoS Biology. 6 2: e35.
- Ratnasingham S, Hebert PDN. 2007. BOLD: The Barcode of Life Data System. Mol Ecol Notes. 7:355–364 (www.barcodinglife.org).
- Reis RE, Kullander SO, Ferraris C. 2003. Check list of freshwater fishes of South and Central America. Porto Alegre, Brazil: Edipucrs.
- Ribeiro AC, Benine RC, Figueiredo CA. 2004. A new species of Creagrutus Günther (Teleostei: Ostariophysi: Characiformes), from Upper Rio Paraná Basin, central Brazil. J Fish Biol. 64:597–611.
- Schaefer SA. 1998. Conflict and resolution impact of new taxa on philogenetic studies of Neotropicalcascudinhos (Siluriformes: Loricariidae). In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS. editors. Phylogeny and classification of Neotropical fishes. Porto Alegre, Brazil: Edipucrs. 375–400.
- Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN. 2005. DNA barcodes reveal cryptic host-specificity within the presumed polyphagus members of a genus of parasitoid flies (Diptera: Tachinidae). Proc Natl Acad Sci U S A. 103 10: 3657–3662.
- Swofford DL. 2002. PAUP*—Phylogenetic analysis using parsimony (*and Other Methods) Version 4.0b10. Sunderland, MA: Sinauer Associates.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA 4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599.
- Templeton AR, Crandall KA, Sing CF. 1992. A cladistic-analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA-sequence data. 3. Cladogramestimation. Genetics. 132:619–633.
- Valdez-Moreno M, Ivanova NV, Elías-Guitiérrez M, Contreras-Balderas S, Hebert PDN. 2009. Probing diversity in freshwater fishes from Mexico and Guatemala with DNA barcodes. J Fish Biol. 74:377–402.
- Vari RP, Harold AS. 2001. Phylogenetic Study of Neotropical Ish Genera Creagrutus Günther and Piabina Reinhardt (Teleostei: Ostariophysi: Characiformes), with a Revision of Cis-Andean Species Smithsonian Contributions to Zoology Number 613. Washington, DC: Smithsonian Institution Press.
- Vari RP, Malabarba LR. 1998. Neotropical ichthyology: An overview. In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS. editors. Phylogeny and classification of Neotropical fishes. Porto Alegre, Brazil: Edipucrs. 1–11.
- Victor BC. 2007. Coryphopteruskuna, a new goby (Perciformes: Gobiidae: Gobinae) from the western Caribbean, with the identification of the late larval stage and an estimate of the pelagic larval duration. Zootaxa. 1526:51–61.
- Ward RD. 2009. DNA barcode divergence among species and genera of birds and fishes. Mol Ecol Resour. 9:1077–1085.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia's fish species. Philos Trans R Soc Lond Ser B Biol Sci. 360:1847–1857.
- Ward RD, Holmes BH, Zemlak TS, Smith PJ. 2007. DNA barcoding discriminates spurdogs of the genus Squalus. In: Last PR, White WT, Pogonoski JJ. editors. Descriptions of new dogfishies of the genus Squalus (Squaloidea: Squalidae). Hobart, Australia: CSIRO Marine and Atmospheric Research. 117–130.
- Ward RD, Holmes BH, Yearsley GK. 2008. DNA barcoding reveals a likely second species of Asian sea bass (barramundi) (Lates calcarifer). J Fish Biol. 72:458–463.
- Witt JDS, Threloff DL, Hebert PDN. 2006. DNA barcoding reveals extraordinary cryptic diversity in an amphipod genus: Implications for desert spring conservation. Mol Ecol. 15:3073–3082.
- Wright S. 1978. Evolution and the genetics of populations 4: Variability within and among natural populations. Chicago, IL: University of Chicago Press.
- Yassin A, Capy P, Madi-Ravazzi L, Ogereau D, David JR. 2008. DNA barcode discovers two cryptic species and two geographical radiations in the invasive drosophilid Zaprionus indianus. Mol Ecol Resour. 8:491–501.
- Yearsley GK, Last PR. 2006. Urolophus kapalensis sp nov., a new stingaree (Myliobatiformes: Urolophidae) off eastern Australia. Zootaxa. 1176:41–52.