Abstract
Background and aims. The application of DNA barcoding as a global standard for fish identification is probing diverse worldwide realms (Nearctic, Australian and the Neotropics) and environments (e.g. marine and freshwater). Comparing the patterns of sequence divergence among conspecific and congeneric taxa between realms can provide valuable information on recent evolutionary histories of lineages as barcode data accumulates. Materials and methods. Herein, we have analyzed over 100 species (around 50%) of the Neotropical fish fauna from the São Francisco River, in southeast Brazil. Our aims were to test the performance of DNA barcoding in this biodiversity-rich region, and to compare patterns of genetic divergence with previous studies. Results. The mean Kimura two-parameter distances within species, genera, families, orders, and classes were 0.5, 10.6, 21.0, 22.7, and 24.4%, respectively, with 100% of the species examined successfully differentiated by barcoding. With the exception of Astyanax bimaculatus lacustris, Piabina argentea, and Bryconamericus stramineus, all other species yield a single, cohesive cluster of barcode sequences. The average ‘nearest-neighbor distance’ was 11.12%, 21-fold higher than the mean within species distance of around 0.54%. In a few instances, deep lineage divergences among conspecifics (up to 10%) and congenerics (up to 22.9%) taxa were revealed. Conclusions. Reflecting possible cases of cryptic speciation and the deeper phylogeographic history of São Francisco fish fauna, with some higher clades extending back into the late Cretaceous and Cenozoic (90 mya), when much of the diversification of the Neotropical region apparently took place. In addition, barcodes also highlighted misidentifications and helped to document range extensions for known species.
Introduction
Neotropical fishes represent 13% of all vertebrates' biodiversity, occurring in less than 0.003% of the world's water (Vari and Malabarba Citation1998). Brazil alone has around 25% of the described freshwater fish species, with over 2587 known species (Buckup et al. Citation2007). However, it is estimated that 30–40% of the Neotropical fish fauna has not yet been described (Reis et al. Citation2003). The Neotropical fish fauna thus offers a challenging group to test the performance of DNA barcoding as universal system for species identification.
The mitochondrial DNA cytochrome c oxidase subunit I gene (COI) has been advocated as a universal tool for the identification of animal species (Hebert et al. Citation2003; Hebert and Gregory Citation2005) and community uptake has been diverse. Applications have included tracking invasive species population sources (Corin et al. Citation2007), wildlife forensics investigations (Dawnay et al. Citation2007; Nelson et al. Citation2007), ecology of cryptic communities (Corin et al. Citation2007; Pfenninger et al. Citation2007), and identification of prey from stomach contents (Pons Citation2006). Reliance on mitochondrial DNA in molecular taxonomy has been criticized because of numerous concerns ranging from introgressive hybridization and pseudogene ontogenesis to retention of ancestral polymorphisms (e.g. Rubinoff Citation2006), all of which could potentially mislead barcoding. However, the examination of species assignment failures typically does not exceed 5–10% (Hebert and Gregory Citation2005; Ward et al. Citation2005; Hubert et al. Citation2008; Valdez-Moreno et al. Citation2009) and indeed are often much lower, suggesting such problems are the exception rather than the norm.
Given the socioeconomic importance of fishes, combined with a solid taxonomic framework against which to validate the performance of barcoding, they were an obvious choice for a large-scale initiative. A global effort to assemble a standardized barcode reference sequence library for all fishes known as FISH-BOL (the Fish Barcode of Life Campaign; Ward et al. Citation2009) was initiated in 2005. Briefly, FISH-BOL aims to highlight cases of range expansion for known species, flag previously overlooked species, facilitate species identification, and enable identification where traditional methods cannot be applied (Ward et al. Citation2009).
Previous barcode studies involving freshwater fishes have detected similar values of sequence divergence within species and among congeners as those involving marine species (Hubert et al. Citation2008; Valdez-Moreno et al. Citation2009). However, the more fragmented environment found in freshwaters when compared with marine habitats is expected to promote a greater genetic structuring among populations and also promote deeper divergences among haplotypes (Ward et al. Citation1994). The lack of strong genetic structure reported in previous studies (Hubert et al. Citation2008; Valdez-Moreno et al. Citation2009) could be due to either a more recent colonization, which occurred after glacial retreats at the end of the Pleistocene (Bernatchez and Wilson Citation1998; Hubert et al. Citation2008), or to a more recent origin of freshwater species (Valdez-Moreno et al. Citation2009).
The São Francisco River basin (SFR), the fourth largest river in Brazil, has a fish fauna of at least 205 species, excluding diadromous species (Alves et al. Citation2007), from which 22 species are considered threatened due to pollution, damming, overfishing, and introduced species (Lins et al. Citation1997). The development of a Barcode database for Brazilian fishes will add new data for the taxonomic identification of Neotropical fishes regardless of lifestage or sex. Also, because there is evidence that cryptic species are prevalent in tropical regions, efforts to document species richness are critical (Hebert et al. Citation2004; Beheregaray and Caccone Citation2007).
We conducted an examination of COI diversity within and among over 100 fish species of the SFR (around 50% of its ichthyofauna) to test whether the divergence of barcode sequences within conspecifics will be lower than within congeneric species, and to evaluate the efficiency of species discrimination by Barcoding. We also test the null hypothesis of a greater genetic structure and divergence among haplotypes when compared with marine species or recent colonized freshwater habitats. As Brazilian freshwater fishes probably have an old evolutionary history (Lundberg et al. Citation1998), a deeper lineage divergence within taxa is expected.
Materials and methods
Samples
Fishes were sampled mainly from the medium course of the SFR, ranging from headwaters to floodplains. Five major tributaries were targeted (Velhas, Paraopeba, Pandeiros, Verde Grande, and Urucuia). Samples were also obtained from one reservoir (Três Marias), small tributaries, and from the main river. In the present study, we did not target marine nor annual fishes. All specimens were photographed and georeferenced. More details on coordinates, collecting localities and dates can be obtained within the project file ‘Brazilian Freshwater Fishes—The São Francisco Basin Population’ (BSB) on the Barcode of Life Data System (BOLD; Ratnasingham and Hebert Citation2007). All specimens have been preserved as reference vouchers at the Museum of Science and Technology, Pontifícia Universidade Católica do Rio Grande do Sul (PUC-RS), and at the fish collection of the Laboratório de Biologia e Genética de Peixes, Universidade Estadual Paulista, UNESP/Botucatu. Morphological identifications were conducted based on the literature, and also by taxonomy specialists at PUC-RS and UNESP collections.
DNA extraction, PCR amplification, and sequencing
Tissue subsamples were isolated from fragments of muscle, fin, and eyes (small voucher species) of 105 fish species from the SFR (one nonindigenous species—Leporinus macrocephalus) and stored in 90% ethanol. Numbers of specimens per species ranged from 1 to 23 with a mean of 4.34 (SD = 2.97). Sequencing was carried out at the Canadian Centre for DNA Barcoding using standard protocols (Hajibabaei et al. Citation2005). An automated proteinase K protocol (Ivanova et al. Citation2006) was used to obtain DNA extracts from prepared tissue subsamples (1 mm3). A fragment of approximately 658 bp COI was amplified using different combinations of primers: FishF1, FishR1 (Ward et al. Citation2005), or the M13-tailed primer cocktails C_Fish F1t1—C_FishR1t1 and C_VF1LFt1—C_VR1LRt1 (Ivanova et al. Citation2007) as noted for each entry in BOLD. The 12.5 μl PCR mixes included 6.25 μl of 10% trehalose, 2 μl ultrapure water, 1.25 μl of 10 × PCR buffer, 0.625 μl MgCl2 (50 mM), 0.125 μl each primer (0.01 mM), 0.0625 μl each dNTP (0.05 mM), 0.0625 μl Taq polymerase (5 U/μl), and 2.0 μl DNA template. Amplification protocols consisted of 94°C for 2 min, 35 cycles of 94°C for 30 s, 52°C for 40 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. The most intense PCR products, visualized on pre-cast agarose gels (E-Gels; Invitrogen, Carlsbad, CA, USA), were selected for sequencing. Sequences were determined bi-directionally using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Inc., Foster City, California, USA) as described before (Hajibabaei et al. Citation2005) using an Applied Biosystems Inc. 3730 or 3130 capillary sequencer.
Data analysis
Sequences were edited and aligned using CodonCode Aligner (v3.5.6; Codon Code Corp., Edham, MA, USA). All data (i.e. electropherograms, trace files, primer details, photographs, and collection localities) for each specimen were deposited within the project ‘Brazilian Freshwater fishes—The São Francisco Basin Population’ file on BOLD (http://www.boldsystems.org) and the assembled DNA sequences were also submitted to GenBank (accession numbers included in Appendix A). Online tools implemented on BOLD were used to estimate pairwise sequences divergences using the Kimura two-parameter (K2P) distance model (Kimura Citation1980), as well as to calculate nearest-neighbor distance (NND) values and neighbor-joining phenograms (for a clear graphic representation of the divergence values, see Appendix B).
Results
A total of 431 COI barcodes were obtained, for 101 species in 75 genera and 22 families, constituting around 50% of all known fauna of the SFR. All amplified sequences were larger than 600 bp, and no insertions, deletions, or stop codons were observed, therefore reducing the possibility of nuclear DNA sequences originating from mitochondrial DNA. Only four species failed to amplify, even after trying different primer combinations: Acestrorhynchus britskii (n = 1), Corydoras garbei (n = 1), Leporinus obtusidens (n = 1), an unidentified genus and species from the family Characidae (n = 4). Accession numbers to BOLD and GenBank sequences for each specimen are provided in Appendix A.
With the exception of two species (Bryconamericus stramineus and Piabina argentea) and one subspecies (Astyanax bimaculatus lacustris), all species analyzed were monophyletic (Appendix B), confirming that barcoding is an efficient method for species-level identification. Moreover, the mean intra-specific distance was less than 1% for 86% of all species (). Mean K2P distances within species, genus, family, order, and class were 0.5, 10.6, 21, 22.7, and 24.4%, respectively (). A 21-fold greater difference among congeneric species than among conspecific specimens was observed. The distributions of mean K2P distances among conspecific individuals and among congeneric species overlapped (K2P distances ranged from 0 to 10.5% among conspecifics and from 0 to 22.9% among congeneric species; ). Deep intra-specific divergences and low sister-species divergence may originate overlapping in the distribution of the genetic distance between conspecifics individuals and congeneric species.
Figure 1. Summary of the genetic variability (K2P distances) distribution at COI sequences for 433 individuals and 101 species. (a) Mean intra-specific distribution of genetic distances. (b) Distribution of the genetic distances to the NN. (c) Pairwise percentage distance to NN versus mean intra-specific distance.
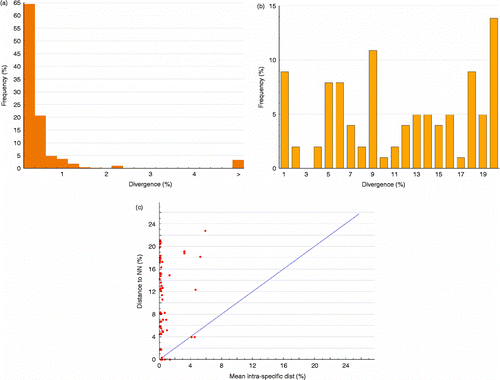
Table I. Summary of genetic divergences (K2P) within species, genus, order, and class.
The lowest congeneric divergences were detected between A. bimaculatus lacustris and A. bimaculatus (0%), Astyanax cf. fasciatus and Astyanax rivularis (0.93%), and also between the commercial important species Prochilodus argenteus versus Prochilodus costatus (1.7%). In addition, the nonidentified specimens Pamphorichthys sp. (BSB361) and Pseudopimelodus sp. (BSB162) had small conspecifics divergence (0–0.15 and 0.33%, respectively), grouping within described species of the same genus. The specimen identified as Brycon sp. (BSB149) was closely related to Oligosarcus sp. (diverging only by 1.8%), possibly due to misidentification, because of their morphological similarity.
On the other hand, deep intra-specific divergences were detected within nine species, with the highest values observed for Pimelodella vittata (). Some of the deep divergences between conspecific could be explained by phylogeographic patterns (e.g. Synbranchus marmoratus and Gymnotus carapo had exclusive lineages detected in the Paraopeba River), reflecting the broad range of sample sites. Other factors such as ancient clade splitting and cryptic speciation might also be related with high genetic intra-specific divergence observed in several small-size species ().
Table III. Species with deep intra-specific barcode divergence.
The NND distribution analysis—that is, the minimum genetic distance between a species and its closest congeneric relative—showed that 9% (nine species) of the NND was lower than 1% ( and ), whereas 89% (90 species) had NND values over 2.7% (). The average NND was 11.12% (ranging from 0 to 25.89%), 21-fold higher than the mean within-species distance of around 0.54, while 88% of intra-specific divergences were less than 1% ().
Table II. Summary of the SFR freshwater fish diversity and distribution of genetic distance.
When considering only the economically important species (large-sized fish [>20 cm], n = 20), all species were monophyletic, with tight clusters (within-species divergences were lower than 1%). Therefore, they were clearly differentiated by barcoding (K2P tree—Appendix C). Only two species, P. argenteus and P. costatus, presented NND lower than 2.7% ().
Discussion
Overall, the observed genetic distances between conspecifics (mean = 0.5%) and congenerics (mean = 10.61%) for the SFR fishes were higher than that previously reported in the marine (0.3 and 8.4%; Ward et al. Citation2009) or freshwater ecosystem (0.3/8.3% and 0.45/5.1%, respectively; Hubert et al. Citation2008; Valdez-Moreno et al. Citation2009). The mean NND value observed herein (11.12%) was also higher than that previously described for the Canadian (7.67%) and Mexican (8.86%) freshwater and Australian marine (9.14%) barcode surveys. Cryptic speciation, phylogeographic structure, and the old evolutionary history of the Neotropical fish fauna, with some higher clades extending back into the late Cretaceous and Cenozoic (90 mya; Lundberg et al. Citation1998) might explain the deep genetic divergence recovered for some SFR species. However, when analyzing commercially important fishes alone (larger species), divergences within species decreased considerably (ranging from 0 to 0.85%, mean = 0.13%) (), possibly reflecting the better taxonomic knowledge of larger species, when compared with small-sized fish.
Deep divergence for congeneric comparisons has been previously reported, with a maximum of 19.3% of divergence (Stoeckle Citation2003; DeSalle et al. Citation2005). Nonetheless, here we report even deeper divergences, with a maximum of 22.9% recovered within the genus Moenkhausia. Interestingly, only 0.1% of NND values were lower than 9% for SFR species ( and ). In contrast, Hubert et al. (Citation2008) reported that 27% of NND values were lower than 0.1%, presumably reflecting the more recent diversification of the freshwater Canadian fauna.
Several cases of possible cryptic speciation due to deep intra-specific barcode divergence were observed, ranging from 2.2 to 10% for several species—e.g. Imparfinis minutus (9%), B. stramineus (9%), Pamphorichthys hollandi (2.2%), P. argentea (6%), and P. vittata (10%)—and well-known species complexes—e.g. Eigenmannia virescens (9%), G. carapo (5%), and S. marmoratus (9%)—are likely to increase genetic divergence estimation (). Biogeography patterns alone are unlikely to explain all deep divergences observed, as pronounced intra-specific genetic divergence was recovered from specimens sampled in sympatricity. For instance, sympatric cryptic lineages diverging up to 10% were recovered from small fish species in the Velhas River Basin—e.g. P. vittata (BSB375-376), B. stramineus (BSB214), and P. argentea (BSB368) ( and Appendix A). Further integrative approaches, including nuclear markers, morphology, and broader range of sampling sites might shed some light on this issue.
Our screen for species splits, applying the threshold of 1% average divergence between conspecific individuals (as suggested by Hubert et al. Citation2008), was able to differentiate most species. Well known as a complex of species of controversial taxonomy (Reis et al. Citation2003), Astyanax specimens were divided into three major clusters, and therefore showed support from barcode data for at least three species: A. bimaculatus, A. rivularis, and A. cf. fasciatus. Even considering the small K2P divergences detected between A. rivularis and A. cf. fasciatus (0.93%), probably indicating a case of recently derived species, a tight array of haplotypes was observed for each cluster (Appendix B). Several nonidentified Astyanax species were also assigned to one of the three clades. However, the subspecies A. bimaculatus lacustris had no divergence from A. bimaculatus; hence, barcodes do not support the hypothesis of divergence between these two subspecific lineages. In another interesting case, samples of P. argentea and B. stramineus could not be separated as expected. Among the seven specimens analyzed, four units were observed. These two genera are composed of several complex species (Javonillo et al. Citation2010; CitationPereira et al. 2010) and new specimens should be analyzed for a clearer picture of this problem, before a resolution could be presented.
Moreover, nine species () would be over-split due to deep divergence within conspecifics if only the 1% threshold was adopted, reinforcing the fact that while divergence thresholds are a useful heuristic tool, they must be applied judiciously. Hubert et al. (Citation2008) reported that the 1% threshold would have overlooked 34 fish species.
Deep intra-specific divergences recovered from S. marmoratus and G. carapo haplotypes might be related to phylogeographic history or geographic structure, as expected for freshwater fishes (Ward et al. Citation1994). Interestingly, both species had lineages exclusive from the Paraopeba River, suggesting a possible Evolutionary Significant Unit (ESU) for this tributary within the SFR. However, due to the few samples analyzed at each site and the lack of additional corroborating data, this result must be considered provisional, but the species are worthy of further phylogeographic investigation. Also P. hollandi, sampled from two different sites (Velhas River and Pandeiros), at least 500 km apart, had 2.1–2.51% within-species divergences. On the other hand, deeply divergent lineages recovered for several species were not always related with phylogeographic patterns. For instance, in I. minutus there was pronounced differentiation (10% sequence divergence) in sympatry (i.e. two lineages found in the Velhas River), which might suggest a possible case of cryptic speciation. Other highly divergent lineages were observed in sympatry: E. virescens had all specimens collected at Pandeiros River with high conspecific lineage divergence (9%) and P. argentea, with 6% of within species divergence, was detected in sympatry with conspecifics in the Curimataí River (tributary of Velhas River Basin). Also, Poecilia sp. presented two clades with 2.2% divergence in sympatry within species samples in the Velhas River. In one case, a young specimen of B. stramineus, which could not be taxonomically differentiated from adults and therefore was identified as B. stramineus, had conspecific divergence of 8.73%, flagging a possible new cryptic species.
Several unidentified small sucker-mouth catfish species (e.g. Bunocephalus sp., Neoplecostomus sp., Pareiorhina sp., Microlepidogaster sp., Parotocinclus sp., Rineloricaria sp., and Hemipsilichthys sp.) were each represented by a single cohesive array of barcode sequences, distinct from any other species, suggesting that each of these genera was represented in our collection by a single species. Interestingly, the specimens identified as Hisonotus sp. are a yet-to-be described genus and species for this river system (Carlos Lucenna personal communication). Other nonidentified small fish species, such as Pamphorichthys sp. (BSB361) and Pseudopimelodus sp. (BSB162), could not be identified based only on morphology, but they were closely associated with known barcoded specimens, of the same genus, with low divergence values (0–0.33%; Appendix B).
Haplotypes of Knodus recovered from SFR species had 100% similarity with Knodus moenkhausii from the Paraná River Basin, as compared with other projects on BOLD, suggesting that these individuals belong to the same species. However, the genus Knodus has not yet been described for the SFR. In addition, Knodus is differentiated from Bryconamericus by a sole feature, the presence of scales on the basal portion of the caudal fin in Knodus (Ferreira and Carvajal Citation2007). We therefore suggest that the range of occurrence of K. moenkhausii should be extended to the SFR. These results are also supported by morphological data (Katiane M. Ferreira, personal communication). Hence, DNA barcodes were able to discriminate Knodus from Bryconamericus (13% of divergence between species), despite little morphological divergence, demonstrating that the barcode approach is valid for Neotropical fish identification. In fact, the species identified here as Knodus were initially misidentified as Bryconamericus; and after our barcode data flagged the likely mistake, their morphological identification was checked and corrected.
In summary, our study provides an example of the usefulness of barcoding for cataloging the diversity of Brazilian freshwater fishes from the SFR. Our barcode data support the discovery of several putative new species and genera (e.g. the ‘Hisonotus sp.’ case), describe a case of range expansion for a known species (e.g. K. moenkhausii) and flagged previously overlooked species (e.g. I. minutus, B. stramineus, P. hollandi, P. argentea, and Poecilia sp.).
We have shown that the current knowledge of the SFR ichthyofauna, even in a well-studied Brazilian river system, is far from complete. The present study was not intended to solve taxonomic issues, but does flag taxa requiring further analyses, as well as providing baseline information on species that may represent good models for comparative phylogeographic surveys (Beheregaray Citation2008). Moreover, a deeper knowledge of the molecular systematics of species complexes of small-sized fishes (which are poorly studied in Brazil) may also help in the description of new species, contributing to improve local species richness estimates and to help delineate taxonomic units for conservation programs. As all commercially important fishes were clearly discriminated, we also foresee a great role of DNA barcode analysis in fish market certification and regulation by governmental agencies.
Supplementary Material
Download MS Word (490.5 KB)Supplementary Material
Download PDF (183.1 KB)Supplementary Material
Download PDF (41.4 KB)Acknowledgements
The authors are grateful to José Vanderval Melo Junior, Arno Soares Seerig, Danilo Pimenta, Bernardo Lage, Ivo, Francisco Andrade, and Lucas Vilea Pires for helping with the collection of fish; to all personnel of the Biology Institute of Ontario and of the Laboratory of Animal Genetics at UFMG Veterinary College, for their assistance; to Luciano Beheregaray for comments on the manuscript; and to Heather Braid for assistance with formatting. The present study was supported by CNPq/FAPEMIG (INCT 573899/2008-8), the Canadian Barcode of Life Network via the Natural Sciences and Engineering Research Council of Canada, the Consortium of Barcode of Life, Instituto Estadual de Florestas and other sponsors listed at www.BOLNET.ca. D.C.C. is also grateful to CNPq for the PDJ fellowship (process number 150420/2009-9).
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Alves CBM, Vieira F, Pompeu PS. 2007. Programa Zoneamento Ecológico-Econômico: Caderno Temático: Biodiversidade. Brasília: MMA/SEDR/SBF110–130.
- Beheregaray LB. 2008. Twenty years of phylogeography: The state of the field and the challenges for the Southern Hemisphere. Mol Ecol. 17:3754–3774.
- Beheregaray LB, Caccone A. 2007. Cryptic biodiversity in a changing world. J Biol. 6:9.
- Bernatchez L, Wilson CC. 1998. Comparative phylogeography of nearctic and palearctic fishes. Mol Ecol. 7:431–452.
- Buckup PA, Menezes A, Ghazzi NA. 2007. Catálogo das espécies de peixes de água doce do Brasil. Rio de Janeiro: Museu Nacional. Vol. 1. 195 pp.
- Corin SE, Lester PJ, Abbott KL, Ritchie PA. 2007. Inferring historical introduction pathways with mitochondrial DNA: The case of introduced Argentine ants (Linepithema humile) into New Zealand. Diversity Distrib. 13:510–518.
- Dawnay N, Ogden R, McEwing R, Carvalho GR, Thorpe RS. 2007. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci Int. 173:1–6.
- DeSalle R, Egan MG, Siddall M. 2005. The unholy trinity: Taxonomy, species delimitation and DNA barcoding. Philos Trans R Soc B Biol Sci. 360:1905–1916.
- Ferreira KM, Carvajal FM. 2007. Knodus shinahota (Characiformes: Characidae) a new species from the Rio Shinahota, Rio Chapare basin (Mamore system), Bolivia. Neotrop Ichthyol. 5:31–36.
- Hajibabaei M, deWaard JR, Ivanova NV, Ratnasingham S, Dooh RT, Kirk SL, Mackie PM, Hebert PD. 2005. Critical factors for assembling a high volume of DNA barcodes. Philos Trans R Soc London B Biol Sci. 360:1959–1967.
- Hebert PDN, Gregory TR. 2005. The promise of DNA barcoding for taxonomy. System Biol. 54:852–859.
- Hebert PDN, Cywinska A, Ball SL, DeWaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc London Ser B Biol Sci. 270:313–321.
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the Neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 101:14812–14817.
- Hubert N, Hanner R, Holm E, Mandrak NE, Taylor E, Burridge M, Watkinson D, Dumont P, Curry A, Bentzen P, Zhang J, April J, Bernatchez L. 2008. Identifying Canadian freshwater fishes through DNA barcodes. PLoS ONE. 3:e2490.
- Ivanova NV, Dewaard JR, Hebert PDN. 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes. 6:998–1002.
- Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. 2007. Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes. 7:544–548.
- Javonillo R, Malabarba LR, Weitzman SH, Burns JR. 2010. Relationships among major lineages of characid fishes (Teleostei: Ostariophysi: Characiformes), based on molecular sequence data. Mol Phylogenet Evol. 54:498–511.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Lins LV, Machado ABM, Costa CNR, Hermann G. 1997. Roteiro metodológico para elaboração de listas de espécies ameaçadas de extinção: Contendo a lista oficial de fauna ameaçada de Minas Gerais. Belo Horizonte: Fundação Biodiversitas.
- Lundberg JG, Marshall LG, Horton B, Malabarba MCSL, Wesselingh F. 1998. The stage for Neotropical fish diversification: A history of tropical South American rivers. In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS. editors. Phylogeny and Classification of Neotropical Fishes. Porto Alegre: EDIPUCRS13–48.
- Nelson LA, Wallman JF, Dowton M, Using COI. 2007. barcodes to identify forensically and medically important blowflies. Med Vet Entomol. 21:44–52.
- Pereira LHG, Pazian MF, Hanner R, Foresti F, Oliveira C. 2010. DNA barcodes discriminate freshwater fishes from the Paraíba do Sul River Basin, São Paulo, Brazil. Mitochondrial DNA 21(S2):1–9.
- Pfenninger M, Nowak C, Kley C, Steinke D, Streit B. 2007. Utility of DNA taxonomy and barcoding for the inference of larval community structure in morphologically cryptic Chironomus (Diptera) species. Mol Ecol. 16:1957–1968.
- Pons J. 2006. DNA-based identification of preys from non-destructive, total DNA extractions of predators using arthropod universal primers. Mol Ecol Notes. 6:623–626.
- Ratnasingham S, Hebert PDN. 2007. BOLD: The barcode of life data system (www.barcodinglife.org). Mol Ecol Notes. 7:355–364.
- Reis R, Kullander SO, Ferraris CJ. 2003. Check List of the Freshwater Fishes of South and Central America. Porto Alegre: EDIPUCRS.
- Rubinoff D. 2006. Utility of mitochondrial DNA barcodes in species conservation. Conserv Biol. 20:1026–1033.
- Stoeckle M. 2003. Taxonomy, DNA, and the barcode of life. Bioscience. 53:796–797.
- Valdez-Moreno M, Ivanova NV, Elias-Gutierrez M, Contreras-Balderas S, Hebert PDN. 2009. Probing diversity in freshwater fishes from Mexico and Guatemala with DNA barcodes. J Fish Biol. 74:377–402.
- Vari RP, Malabarba LR. 1998. Neotropical ichthyology: An overview. In: Malabarba LR, Reis RE, Vari RP, Lucena ZMS, Lucena CAS. editors. Phylogeny and Classification of Neotropical FishesPorto Alegre1–11.
- Ward RD, Woodwark M, Skibinski DOF. 1994. A comparison of genetic diversity levels in marine, freshwater, and anadromous fishes. J Fish Biol. 44:213–232.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia's fish species. Philos Trans R Soc London B Biol Sci. 360:1847–1857.
- Ward RD, Hanner R, Hebert PDN. 2009. The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol. 74:329–356.
