?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
We prepared multifunctional envelope-type nano device (MEND) loading siRNA through lipid film hydration method; the qualities of MEND were characterized by the shape, particle size, and evaluated by the encapsulation efficiency (EE), plasma stability, and transfection efficiency. The formulated MEND was found to be relatively uniform in size with a positive zeta potential. The average siRNA entrapment efficiency was 86.4%. After 48 h exposure in 50% (v/v) serum at 37°C, the siRNA in MEND had no significant degradation. When the concentration of siRNA in 24-well plates was 500 nM, the MEND had high transfection efficiency, almost 100%.
Introduction
RNA interference (RNAi) is a newly developed technology (CitationBaigude et al. 2013), and has been applied in the therapy of tumor, viral infection, hepatitis B, and many other diseases. siRNA is the effector molecule which induces the RNAi in vivo. But naked siRNA is easily degraded by RNases in vivo, and the half-life is short (CitationShi et al. 2009), and too large (˜13 kDa) and too negatively charged to cross cellular membranes (CitationWhitehead et al. 2009). Regarding the reasons above all, the naked siRNA needs the help of vectors to improve its circulation time, penetrate the cell membrane, and take action.
For the efficient delivery of siRNA to the cells, an ideal non-viral vector must have various functions. We succeeded in constructing a delivery system which is called multifunctional envelope-type nano device (MEND) by a lipid film hydration method based on electrostatic interactions between each of the components, such as siRNA, polycations (Poly-L-lysine, PLL), and functional lipids (MEND/CHEMS). The MEND modified with PLL was positively charged at physiological pH 7.4, and was found to enhance transfection activities compared with MEND without PLL modifying in HepG2 cells.
DOPE/CHEMS as the pH-sensitive fusogenic lipid, have the characteristics of pH-sensitive liposomes. They are designed to be stable at physiological pH 7.4 but to undergo destabilization and acquire fusogenic properties under acidic conditions, thus leading to the release of their encapsulated siRNA. Endosomes and tumor exhibit an acidic environment as compared to normal tissues, so the packaging of siRNA-PLL with pH-sensitive fusogenic lipid (DOPE/CHEMS) improved the transfection activity, passively due to their enhanced endosomal escape (CitationKim et al. 2008).
Materials
Instruments
Electrophoresis instrument (DYY-8C), automatic digital gel imaging analysis system (JS-380B), bath ultrasonic cleaner (SK5210HP), Centrifuge (D-78532), Centrifugal filter devices (Amicon®Ultra-4, Millipore), Nuclepore Track-Etch Membrane (Whatman®), fluorescence microplate reader (SoftMax®Pro5), Zetasizer Nano-ZS90, transmission electron microscopy (TEM) (JME2010 TEM, JEOL), and inverted fluorescence microscope (Axio Observer A1) were used.
Reagents
NC-siRNA and FAM-siRNA were synthesized by Gene Pharma Co., Ltd (Shanghai, China). The reagents included PLL (MW30.000 ˜70.000, Sigma), agarose9 (G-10, Spain Biowest), 1, 2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE, Genzyme, corden), 3β-hydroxy-5-cholestene3-hemisuccinate (CHEMS, Avanti), fetal bovine serum (FBS) (Hyclone® Thermo Fisher Scientific), RPMI-1640 (Hyclone® Thermo Fisher Scientific), and 0.25% Trypsin solution (Hyclone® Thermo Fisher Scientific). Other materials were all analytical or pharmaceutical grade.
Cell culture
HepG2 cells were cultured in cell-culture flasks (Denmark NUNC Company) containing RPMI-1640 supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 mg/ml) at 37°C in an atmosphere of 5% CO2 and 95% humidity.
Methods
Preparation of PLL-siRNA complexes and Gel retardation assay
The PLL-siRNA complex was obtained by adding siRNA to diethy pyrocarbonate treated water (DEPC) solution with different concentrations of PLL. Their mass ratios were 1, 3, 5, 8, 10, 15, 30, 40, and 50, respectively. The formation and charge properties of PLL-siRNA complex were confirmed by Gel retardation assay. The electrophoresis buffer's pH was 7.4.
Preparation of MEND
Operating instructions
Procedure for the preparation of MEND comprised three steps as follows.
(i) siRNA condensation with PLL: siRNA and PLL were dissolved with DEPC-treated water. To condense the siRNA, the siRNA solution (20 μM) was mixed with a PLL solution (2.7 mg/ml) under vortexing at room temperature. The siRNA content of suspension of siRNA/PLL complex was prepared at mass ratio of 10. (ⅱ) Hydration of the lipid film: After the condensation of siRNA, siRNA-PLL suspension was added to the lipid film, formed by the evaporation of a chloroform solution of lipids, [DOPE/CHEMS = 6:4 (molar ratio)] on the bottom of a round-bottomed flask, followed by incubation for 15 min to hydrate lipids. (ⅲ) Sonication for the packaging of the condensed siRNA: To coat siRNA-PLL with lipids, the round-bottomed flask was then sonicated for about 3 min in a bath-type sonicator. Furthermore, to attach the positively charged peptide PLL to the envelope of the lipid-coated particle, an PLL solution (60 μl, 2.7 mg/ml) was added to the suspension, and the mixture was incubated for 30 min at room temperature (CitationKogure et al. 2004). The resulting MEND suspension was made uniform by passing through nucleopore membranes of 200 and 80 nm for three times respectively (CitationGao et al. 2011).
Evaluation of siRNA encapsulation efficiency (CitationGao et al. 2012)
The siRNA encapsulation efficiency (EE) was determined by ultra-filtrating the liposomes entrapping FAM-siRNA using Amicon®Ultra-4 centrifugal filter devices (10,000 NMWL, Millipore). After thorough ultra filtration with DEPC water, unencapsulated FAM-siRNA was collected and quantified using a calibration line obtained with standard lead FAM-siRNA solutions. The fluorescence of FAM-siRNA was determined with the fluorescence microplate reader using excitation and emission wavelengths of 483 and 525 nm, respectively. The siRNA EE was calculated using the formula:
Mu and Mi were defined as the mass of unencapsulated siRNA and initially added siRNA, respectively.
The preparation optimization of MEND (CitationDu et al. 2010, CitationAn et al. 2011)
The EE was the indicator of the optimization study on the preparation of MEND. The factors, including liposome concentrations, the mole ratio of DOPE and CHEMS, the dosage of siRNA, the pH of PBS, the volume of rehydration solution, temperature and rotation speed, were considered to optimize the main influencing factors which affect the preparation of MEND. Integrating the results of the single factor investigations, the principal factors were selected for the orthogonal tests of the preparation of MEND.
Characterization of MEND
Shapes of MEND
The MEND was dropped in a microscopic copper grid, negatively stained with 0.03% phosphotungstic acid (PTA) and dried, then the stained sample was observed through the TEM (CitationAn et al. 2011, CitationZhou et al. 2010).
Particle size and zeta potential of MEND
We evaluated the average size and zeta potential of MEND (non-PLL), MEND (PLL) by using Zetasizer Nano instrument. The original solution of MEND (non-PLL) and MEND (PLL) were diluted seven times, respectively.
EE of MEND
The experiments of orthogonal tests indicated that the best combination of factors and levels was A1B2C3D1. According to the optimal prescription, MEND was prepared and EE of MEND was determined by the fluorescence microplate reader.
siRNA serum stability
Serum stability of siRNA in its aqueous solution, PLL complexes, and liposome formulations was evaluated using agarose gel electrophoresis. Samples of siRNA either in aqueous solution, PLL complexes, or liposomes were mixed with FBS in a 1:1 volume ratio to give 50% serum concentration and incubated at 37°C. At different times, aliquots containing 0.25 mg siRNA of each sample (the liposomes were first destroyed by Triton X-100) were loaded onto a gel and electrophoresis was performed to visualize intact siRNA.
In vitro cellular uptake (CitationGao et al. 2012)
For in vitro cellular uptake analysis, HepG2 cells were seeded in 24-well plates at a density of 4 × 104 cells per well about 20 h. Cells were treated with MEND (PLL) and pure siRNA at a concentration of 250 nM or 500 nM FAM-siRNA for 10 h. After that, cells were washed two times with PBS. Fluorescence microscope was employed to examine the intracellular distribution of siRNA after cellular uptake.
Quantitative determination of siRNA uptaken by the HepG2 cells was performed using a fluorescence microplate reader. Briefly, HepG2 cells were seeded in 24-well plates at a density of 4 × 104 cells per well for about 20 h. Cells were treated with the liposomes at a concentration of 500 nM FAM-siRNA and with pure FAM-siRNA at a concentration of 500 nM for 10 h. After that, cells were washed three times with PBS followed by incubation with 300 ml lysis buffer (5% Triton X-100 in PBS) at RT for 0.5 h. The quantity of FAM-siRNA in the cell lysis solution was quantified using a calibration line obtained with standard FAM-siRNA solutions. The fluorescence of FAM-siRNA was determined with fluorescence microplate reader using excitation and emission wavelengths of 483 and 525 nm, respectively. The siRNA uptake percentage was calculated from the following formula:
QU and QI were defined as the mass of uptaken siRNA and initially added siRNA, respectively.
Results
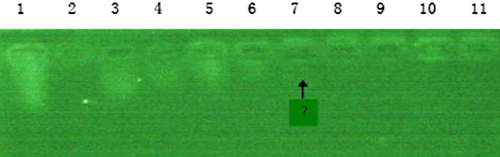
Gel retardation assay of siRNA/PLL complex
The result was shown in . With increase in the mass ratio of PLL and siRNA, the siRNA swimming from the sample hole became less. When the mass ratio of PLL to siRNA is 10 (lane7), siRNA-PLL was composed completely of PLL.
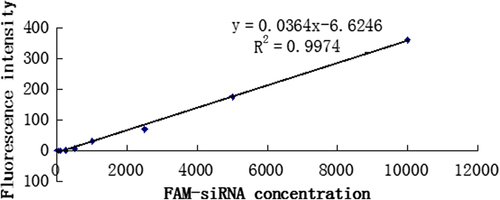
Liner relation of FAM-siRNA concentration and fluorescence intensity
The siRNA EE was calculated using the formula:
Mu and Mi were defined as the mass of unencapsulated siRNA and initially added siRNA, respectively. FAM-siRNA concentration and fluorescence intensity had a good linearity with FAM-siRNA concentrations ranging from 10 ng/ml to 10 μg/ml, Y = 0.0364X − 6.6246, R2 = 0.9974. The standard curve was shown in .
The preparation optimization of MEND
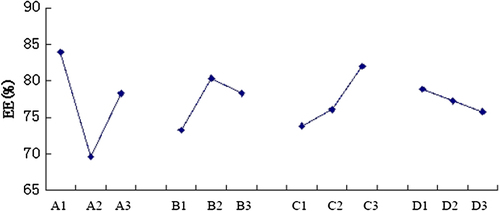
Integrated the results of the single factor investigations, the factor A (the mole ratio of DOPE and CHEMS), factor B (the dosage of siRNA), factor C (the pH of PBS), and factor D (the volume of rehydration solution) were selected as the principal factors of the orthogonal tests of the preparation of MEND ( and ), each factor has three levels, to prepare MEND at 45°C.When the EE of MEND was treated as the indictor, the intensity orders of the factors which affect the EE of MEND was A > C > B > D.
Table I. Factors and levels of orthogonal tests.
Table II. Arrangement and results of orthogonal tests according to L9(34).
As was shown in , the EE of MEND was largest at level 1 of factor A, level 2 of factor B, level 3 of factor C, and level 1 of factor D, respectively. The result of optimal combination was A1B2C3D1, the conditions were 6:5 (DOPE/CHEMS mole ratio), 10 μg siRNA, pH 6.0, and 1 ml PBS.
Data analysis
The principal factor depended on the R (range) value, the factor sequence that influenced the EE was A > C > B > D, the best combination of factors and levels was A1B2C3D1.
Characterization of MEND
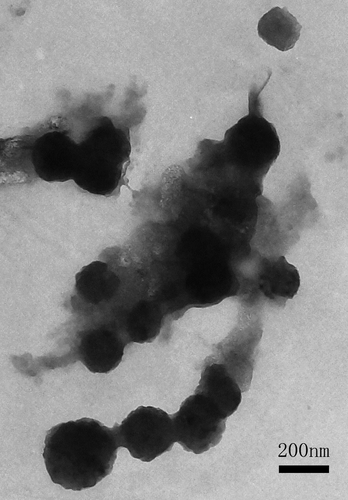
Shapes of MEND
TEM observations revealed that MEND had been successfully produced and the shape of the liposome was round with estimated diameter at 170 nm ().
Particle size, zeta potential, and EE of MEND
The size and zeta potential of MEND (non-PLL) as determined were about 130 nm with negative charge, and MEND (PLL) as determined were about 170 nm with positive charge (). As is shown in , MEND (PLL)'s average size was larger than MEND (non-PLL), and their charges were opposite.
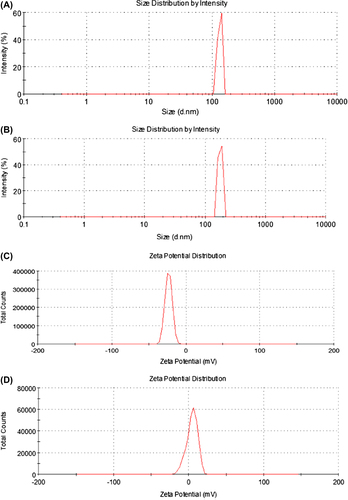
Table III. Particle size and of zeta potential MEND.
A1 is the size distribution of MEND (non-PLL), A2 is zeta potential distribution of MEND (non-PLL); B1 is the size distribution of MEND (PLL), B2 is zeta potential distribution of MEND (PLL).
When the MEND was prepared in accordance with the optimal formulation, the EE was 86.40 ± 1.03%.
siRNA serum stability
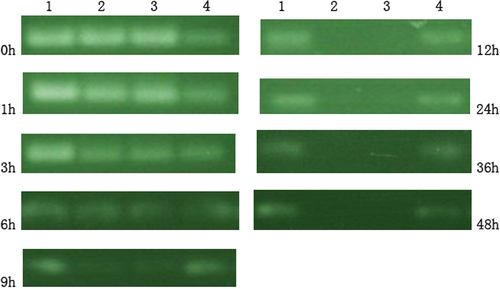
As shown in , lane 1 was marker, lane 2 was pure siRNA, lane 3 was siRNA-PLL, and lane 4 was MEND. The amount of siRNA at lane 2 was 35.4% of the marker, lane 3 was 38.4% of the marker, and lane 4 was 97.7% of the marker at 3 h; the amount of siRNA at lane 2 was 10.3% of the marker, lane 3 was 11.2% of the marker, and lane 4 was 89.2% of the marker at 9 h; siRNA in lane 2 and lane 3 was degraded absolutely and siRNA in lane 4 was 86.4% of the marker at 12 h; at 48 h, the amount of siRNA was 58.2%.
In vitro cellular uptake
As demonstrated by fluorescence microscope analysis, MEND had a high transfection efficiency (almost 100%) in HepG2. Pure siRNA had a low transfection efficiency, and the fluorescence intensity in cells was so weak that fluorescence microscopy can't detect the intensity. The fluorescence intensity in HepG2 cells which were treated with the liposomes entrapping 500 nM siRNA was stronger than the fluorescence intensity in HepG2 cells which were treated with the liposomes entrapping 250 nM siRNA () where (A) MEND (PLL) at a concentration of 500 nM FAM-siRNA. (B) MEND (PLL) at a concentration of 250 nM FAM-siRNA. (C) pure FAM-siRNA at a concentration of 500 nM. (D) pure FAM-siRNA at a concentration of 250 nM.
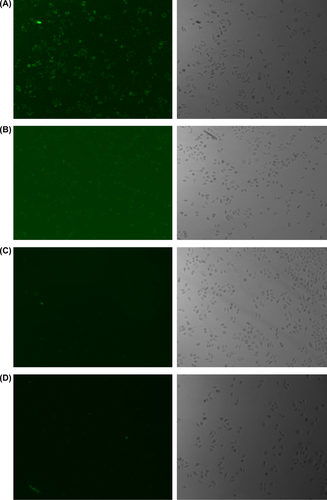
Considering fluorescence microscope analysis can only reflect the relative value of transfection efficiency, we further employed a fluorescence microplate reader to determine the actual quantity of FAM-siRNA uptaken. Consistent with the fluorescence microscope analysis results, MEND (PLL) showed significantly higher siRNA uptake percentage compared with pure siRNA at a concentration of 500 nM FAM-siRNA. At 10-h time point, MEND (PLL) showed a very high siRNA uptake percentage (44.06%), while pure siRNA only had a very low siRNA uptake percentage (3.25%). Taken together, these data suggested that the in vitro cellular uptake of MEND (PLL) was significantly higher than that of pure siRNA.
Discussions
Because of the poor stability of siRNA, in the process of preparation of MEND, siRNA should be protected well. Some measures should be taken, such as avoiding high heat, long and high-power ultrasound, and strong acid and alkali environment. DEPC water must be used as the solution to protect siRNA from RNase.
Many scientists made positive liposomes usually by the following way. First, using lipid film hydration method blank liposomes were made. Then siRNA solution was poured into blank liposomes and both were mixed. By this way, we found that siRNA was mostly absorbed at the surface of blank liposomes. To enhance the EE and stability of siRNA, siRNA solution should be added at the hydration time. We found that siRNA was coated with the lipid bilayer.
Results of preparation optimization experiments of MEND indicated that when the MEND was prepared in accordance with the optimal formulation, the EE was large. At pH 7.4, PLL-siRNA complex's charge was weak, as electric charge of PLL could just neutralize the charge of siRNA. So at pH 6.0, PLL-siRNA complex was positive charge, DOPE/CHEMS as the anionic liposome materials was negative charge. Due to the strong electrostatic interactions of PLL-siRNA and liposome, PLL-siRNA complex was almost entirely encapsulated. But the pH could not be reduced, as DOPE/CHEMS is pH-sensitive fusogenic lipid, when pH was lower than 6.0, liposome underwent destabilization and acquired fusogenic properties under acidic conditions. In the premise of ensuring a high EE, the pH could not be reduced below 6.0.
Serum stability assay showed that pure siRNA and siRNA-PLL started to degrade after 3 h and was fully degraded after 9 h. But siRNA in MEND did not clearly degrade even after 36 h. The results suggested that PLL's role in protection of siRNA degradation in serum was not obvious. However, MEND could protect siRNA from serum degradation to a great extent, suggesting that the functional lipids provide a favorable protection to siRNA degradation in serum. We speculated that PLL was just mixed with siRNA, however, lipid film could have wrapped the siRNA. Because of the isolation of lipid film, the degradation of siRNA was slight.
At transfection experiments, as the cell surface is negatively charged, so the positively charged MEND (PLL) could enrich around tumor cells by electrostatic interactions. The higher the concentration of the MEND (PLL) around cells, the more siRNA could be transfected into the HepG2 cells. Due to the electrostatic repulsion between negatively charged MEND (CHEMS), pure siRNA, and negatively charged cells, the transfection efficiency of siRNA in HepG2 was relatively low (CitationObata et al. 2008).
All results indicate that we have successfully prepared MEND which has the nanoparticles and liposomes’ characteristics. Also, MEND can incorporate various functional devices such as a specific ligand to specific cells, intracellular sorting devices that permit endosomal escape, long circulation, etc. In the near future, we will use anti-luciferase siRNA to perform transfection experiments and modify MEND with functional devices.
Declaration of interest
The authors report no declarations of interest. The authors alone are responsible for the content and writing of the paper.
This work was supported by the National Natural Science Foundation in 2014 (81373350) and Qingdao Science & Technology project in 2013 (13-1-3-49-nsh).
References
- An K, Sun Y, Xu L, Cui X. 2011. Preparation and in vitro evaluation of simvastatin ethosome. Artif Cells Blood Substit Immobil Biotechnol. 39:347–350.
- Baigude H, Su J, McCarroll J, Rana TM. 2013. In vivo delivery of RNAi by reducible interfering nanoparticles (iNOPs). ACS Med Chem Lett. 4:720–723.
- Du B, Li Y, Li X, A Y, Chen C, Zhang Z. 2010. Preparation, characterization and in vivo evaluation of 2-methoxyestradiol-loaded liposomes. Int J Pharm. 384:140–147.
- Gao J, Liu W, Xia Y, Li W, Sun J, Chen H, et al. 2011. The promotion of siRNA delivery to breast cancer overexpressing epidermal growth factor receptor through anti-EGFR antibody conjugation by immunoliposomes. Biomaterials. 32:3459–3470.
- Gao J, Yu Y, Zhang Y, Song J, Chen H, Li W, et al. 2012. EGFR-specific PEGylated immunoliposomes for active siRNA delivery in hepatocellular carcinoma. Biomaterials. 33:270–282.
- Kim MJ, Lee HJ, Lee IA, Kim IY, Lim SK, Cho HA, Kim JS. 2008. Preparation of pH-sensitive, long-circulating and EGFR-targeted immunoliposomes. Arch Pharm Res. 31:539–546.
- Kogure K, Moriguchi R, Sasaki K, Ueno M, Futaki S, Harashima H. 2004. Development of a non-viral multifunctional envelope-type nano device by a novel lipid film hydration method. J Control Release. 98:317–323.
- Obata Y, Suzuki D, Takeoka S. 2008. Evaluation of cationic assemblies constructed with amino acid based lipids for plasmid DNA delivery. Bioconjug Chem. 19:1055–1063.
- Shi ML, Zhao ZH, Wang Y, Chen HP. 2009. In vivo delivery of siRNA. Herditas. 31: 683–688.
- Whitehead KA, Langer R, Anderson DG. 2009. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 8:129–138.
- Zhou Y, Wei YH, Zhang GQ, Wu XA. 2010. Synergistic penetration of ethosomes and lipophilic prodrug on the transdermal delivery of acyclovir. Arch Pharm Res. 33:567–574.