Abstract
The aim of this study was to design a targeted drug delivery system carrying a natural anticancer drug Quercetin (Qu), specifically for skin cancer. A central composite design was applied separately for each ligand, and the quadratic model was used for the process. The surface morphology was confirmed by transmission electron microscopy (TEM) and Fourier transform infrared spectroscopy (FTIR), and in vitro release studies were also performed. The MTT assay was performed against two different cell lines, to measure their anticancer potentials and their targeting ability. The study thus reveals that MA-Qu-PLGA and FA-Qu-PLGA nanoparticles (NPs) can be used as effective drug delivery systems for skin cancer treatment encompassing natural drugs.
| Abbreviations | ||
| EDC | = | N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide |
| NHS | = | N-hydroxysuccinimide |
| PLGA | = | Poly (lactide-co-glycolide) |
| PBS | = | Phosphate-buffered saline |
| DCM | = | Dichloro methane |
| DMSO | = | Dimethyl sulfoxide |
| PA | = | physical adsorption |
| TEM | = | transmission electron microscopy |
| DLS | = | dynamic light scattering |
| MA-Qu-PLGA | = | Mannose-Quercetin- Poly lactide-co-glycolide |
| FA-Qu-PLGA | = | Folate-Quercetin-Poly lactide-co-glycolide |
| EE | = | Entrapment Efficiency |
| C.V | = | coefficient of variation |
| NP | = | Nanoparticle |
Introduction
Skin cancer is a disease which has acquired numerous forms over the years. Several drug delivery systems have been developed, to deliver the therapeutic drug at the desired site, but due to constant wear and tear of the keratinized layer of the skin, the lipophilicity of membrane, the presence of enzyme systems, and many other contributing factors, drug delivery to the skin is not as easy as it seems to be. A targeted drug delivery system, employing the method of active targeting by conjugating the ligands that are specifically recognized by surface receptors of target cells, can overcome this problem (Soppimath et al. 2001). To contribute to this aspect of active targeting, NPs embrace unique characteristics in terms of appearance and application. Presently, a huge range of synthetic and herbal drugs, biological enzymes, small hydrophilic and hydrophobic drugs, vaccines, and macromolecules can be loaded or encapsulated in the NPs, for effective delivery (Makadia and Siegel 2011, Muller et al. 2007, Wissing et al. 2002, Schafer-Korting et al. 2007).
Functionalization of these NPs by either conjugating or binding them with appropriate ligands can prove to be a better option to treat skin cancer synergistically at different layers.
Currently, there are a variety of ligands that are used to activate these NPs for the purpose of targeting, and include proteins, peptides, antibodies, polysaccharides, glycolipids, glycoproteins, and lectins (Uner 2006, Han et al. 2008, Lin et al. 2007, Lee et al. 2002).
The aim of our study was to utilize two of these ligands for targeted drug delivery of a natural anticancer drug. D(+) Mannose is ligand-specific for the mannose (MA) receptor, which is an endocytic receptor present on the surface of macrophages, and is reported to be extensively involved in the growth of ultra violet radiation-induced skin carcinoma (Wang et al. 2008, Mittal et al. 2010). Similarly, the folate (FA) receptor has an affinity for folate, and is represented on a number of cancerous cells. The reasons behind selecting these receptors and their corresponding ligands are:
Both the receptors are present on the cell surface of cancerous cells, and can recognize their specific ligand-conjugated drug moieties (mannosylated conjugates and folate conjugates).
They both carry out clathrin-dependent endocytosis.
The receptors have an affinity towards those ligands which are biocompatible and physiologically inert.
Among all other receptors, these are the best suited for the design of a drug delivery system, especially for small molecules and macromolecules, and are ideal for natural bioactives ().
With the process point of view, these ligands and their formulation are sufficiently cheap and affordable, to use for further development.
Experimental: Materials and methods
Materials
Quercetin (Qu) was purchased from Sigma Aldrich, USA; Poly(lactide-co-glycolide) copolymer (PLGA) (Mw∼17000–22000, 50/50) was obtained with thanks from Resomer, Evoniks, Germany; Folate (FA), N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide (EDC), and N-hydroxysuccinimide (NHS), were purchased from Sigma-Aldrich, India, and D(+) Mannose (MA) was purchased from HiMedia Laboratories. Phosphate-buffered saline (PBS) (pH 5.5 and pH 5.6) was used for drug release. All other reagents used were of analytical grade.
Chemicals for cell culture
Qu was purchased from Sigma, USA. All solvents were of HPLC grade. Water was purified using a Millipore Super Q water system. The two cell lines were obtained from the National Center for Cell Science, Pune, India. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was purchased from Sigma Aldrich Ltd. (India). HaCaT cells were cultured in Dulbecco's Modified Eagle's Medium Nutrient Mixture F-12 HAM (DMEM F-12 HAM), with 2 mM L-glutamine supplemented with 10% fetal bovine serum (FBS), 45 IU/ml penicillin, and 45 IU/ml streptomycin (HaCaT media), at 37°C in 5% CO2.
Synthesis of Qu-PLGA NPs
The Qu-encapsulating PLGA NPs were prepared as per the method described by Fessi et al. (2002), with slight modification. Briefly, 50 mg of PLGA and 20 mg of Qu were dissolved in about one ml of acetone, and then the solution was directly injected into 4 ml of Millipore-distilled water under sonication. An additional volume of water was added and further sonicated for 20 min, to obtain homogeneous nanocolloids (Stella et al. 2000, Narayanan et al. 2010, Nobs et al. 2004, Nukolova et al. 2011). The organic solution was removed from the sample by evaporating it in a Rota evaporator (IKA ® RB 10) at 37°C, under reduced pressure. The Qu-encapsulated PLGA NPs were separated from the un-entrapped Qu by centrifugation (Remi, C-24 BC) at 15,000 rpm for 30 min. The pellet was re-suspended in MilliQ-purified water after repeated centrifugation, and further subjected to lyophilization in the lyophilizer (Thermo Fisher, Germany & Thermo Fisher Heto LL 3000). The lyophilized sample was sealed and stored at 4°C for further analysis (Lu et al. 2012, Zhang et al. 2012, Dosio et al. 2010).
Bioconjugation of Qu-PLGA NP with folate
The conjugation of folate (FA) with the Qu-PLGA NP was done by activating the carboxylic end of the Qu-entrapped PLGA NP by EDC–NHS conjugation chemistry. Qu-PLGA NPs (1 mg ml− 1) were suspended in PBS buffer (pH 5.6) and incubated in the dark with EDC for 30 min, at room temperature. Immediately, the sample was mixed with NHS for 6 h, under parallel conditions. The sample was then washed several times with Millipore water and filtered. (Dosio et al. 2010, Xia et al. 2008, Hentschel et al. 2008) To the filtrate, FA was added at a concentration of 100 μgml− 1 in PBS, at a pH of 5.5, under the condition of overnight stirring. The filtrate was again centrifuged for 20 min at 12,000 rpm. Excess linking reagent and soluble byproducts were removed by washing thrice with 1 ml of PBS, at a pH of 5.5. Finally, the FA-NP-Qu was lyophilized further to get the dry sample.
Attachment of the Qu-PLGA nanoparticle with MA
Attachment of MA to the NP was done as per the method suggested by earlier researchers, with slight modifications. (Wu et al. 2012) Briefly, about 20 mg of MA was added to 2 mg of Qu-PLGA NPs dispersed in acidic buffer (pH 5.5), and again kept under stirring for 48 h. The sample was then collected, filtered, washed, and centrifuged (Remi, C-24 BC) at 10,000 rpm for 20 min at 4°C. The final product was further lyophilized to get the dried sample. Further, the MA-Qu-PLGA NPs were characterized by FTIR spectroscopy.
Physico-chemical characterization of ligand-conjugated Q-PLGA NP
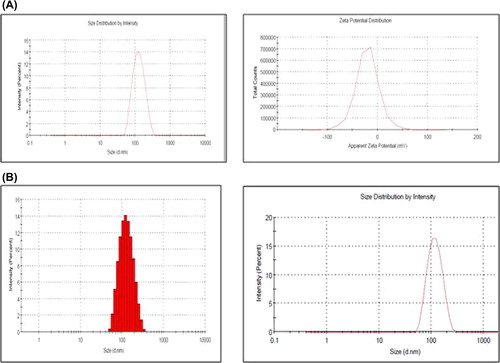
The average size and size distribution of the drug-loaded NPs were measured by the dynamic light scattering (DLS) technique, using a particle size analyzer (Malvern®, Zetasizer, ZF-90) (Teeranachaideekul et al. 2007, Mehnert et al. 2001, Abu-Izza et al. 1996, Kassama et al. 2008). Samples for measurement were prepared by diluting the material suspension with MilliQ water. The hydrodynamic particle size was determined by DLS, which measures the intensity of light scattered by particles in the sample. TEM was employed to study the morphology as well as particle size of the ligand-conjugated Qu NPs. The zeta potential, a measure of the stability of colloidal suspensions, was obtained by measuring the mobility distribution of a dispersion of charged particles as they were subjected to an electric field. The zeta potential was also measured at the same dilution and temperature, to examine the surface charge as well as stability of the NPs (Huang et al. 2004, Ahn et al. 2008, Zhang et al. 2010). FTIR was used to identify the infrared absorption peaks of Qu, PLGA, FA, MA, MA-Qu-PLGA, and FA-Qu-PLGA NPs, to confirm the conjugation. The spectral scan was carried out in the frequency range spanning 4000 to 400 cm− 1 using a Shimadzu FTIR- 8400S. As per the standard procedure, 2 mg of the material to be tested was mixed with 175 mg of KBr, following which it was pelletized for IR measurements. All the measurements were done in triplicate. X-ray diffraction (XRD) patterns of physical mixtures of Qu/PLGA/FA, and physical mixtures of MA/PLGA/Qu, and their lyophilized NPs (≈1gm) were recorded using an XRD instrument (Bruker AXS D8 Advance, India, and Temperature Attachment unit Anton Paar, TTK 450) employing a cobalt (Co) tube at λ = 1.78899 A, running at 30 kV and 16mA. ()
Being a natural flavonoid, Qu was found to have a poor aqueous solubility, but was found to be soluble in most of the organic solvents. The partition coefficient of free Qu was found to be 2.96, which confirms the lipophilicity of the drug, and hence the nanoparticulate delivery of the drug provides an option for the delivery of hydrophobic drugs. The solubility of Qu, Qu-NPs, and ligand-conjugated Qu-NPs were tested in various common solvents. Briefly, a definite quantity (10 mg) of drug was dissolved in 10 ml of each of the solvents investigated, at room temperature. The solubility was observed by the visual inspection. The NPs were completely dispersed in the aqueous medium, which was not observed with free Qu.
Entrapment efficiency (%) determination
For the evaluation of drug entrapment efficiency (EE), the NPs were centrifuged at 15000 rpm for 30 min, and then the supernatant was analyzed using the UV-VIS absorption spectrometer (UV-1700 Pharma Spec, SHIMADZU) at 275 nm, taking the reference of the standard curve plotted for various concentrations of Qu in phosphate-buffered saline PBS (pH 5.5) (Liu et al. 2010). Further, based on the standard curve of Qu, the concentration of the drug in the supernatant was deciphered, and the EE was deduced using the following formula:
Where “MNP” is the mass of drug in the NPs and “Mformulation” is the mass of drug used in the formulation.
The in vitro release of Qu from the ligand-conjugated PLGA NPs was performed at 37°C in phosphate-buffered saline (PBS). Lyophilized NPs of FA-Qu-PLGA were suspended into Eppendorf Tubes containing PBS (1/10 w/v) at pH levels of 5.5 and 5.6, and incubated at 37° C under magnetic stirring (Gambhire et al. 2011, Ghasemian et al. 2013, Chidambaram et al. 2011, Cooper et al. 2014, Yadav et al. 2010). At determined time intervals, the tubes were centrifuged at 15,000 rpm for 15 min, and the supernatants were spectrophotometrically analyzed. The standard curve for Qu was used to determine the amount of the drug released at different time intervals (Ahlin et al. 2002). All the measurements were done in triplicate.
Determination of folate content on the surface of NPs
The amount of folate present on the surface of NPs prepared by the dialysis method was determined using a UV spectrophotometer. The analysis was carried out in CH2Cl2/DMSO (1/4) solvent. The NPs were evaluated by measuring the absorbance of the sample at 358 nm (folate ε = 15,760 M− 1 cm− 1). All the measurements were done in triplicate (Gambhire et al. 2011).
Determination of MA content on the surface of the NPs (phenol sulfuric acid reaction method)
For the quantification of MA, a modified phenol sulfuric acid reaction method was employed. Briefly, 2 mg of NP was dissolved in 200 μL of DCM in an Eppendorf Tube®, and 500 μL of water was then added to this. The two separate phases were mixed thoroughly. The DCM was separated by heating upto 50°C, and the aqueous phase was separated. The sample was then centrifuged at 15,000 rpm for 15 min at low temperature. To remove the insoluble fraction, the sample was collected and washed at least three times (Wu et al. 2012). The supernatant and the washed samples were analyzed by colorimetric assay, in which the supernatant was mixed with concentrated sulfuric acid (96%) and phenol solution, incubated for few minutes at high temperature, and further analyzed for color development by cooling to room temperature. The absorbance was measured at 490 nm, using a microplate reader. The amount of MA incorporated in NP was calculated using the formula:
In vitro drug release
The release of Qu from the MA-conjugated as well as the FA-conjugated PLGA NPs, prepared by the modified PA and EDC-NHS methods respectively, were determined at two pH conditions, that is, pH 5.5, to trigger the release of macrophages in the endosomal compartment, and pH 5.6, to match the physiologic conditions of the skin.
MTT Assay
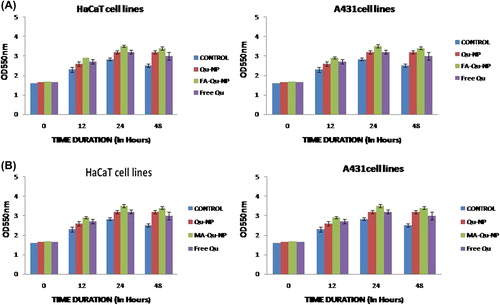
Cells were seeded in 96-well plates at a density of 1 × 105 cells/well. Thereafter, cells of both the cell lines (HaCaT & A431) were subjected to DMSO and DMSO containing FA-Qu-NP and MA-Qu-NP, at concentrations ranging from 0 to 75 μm for both the cell lines, taken as control and test samples respectively. The cells were treated for 48 h. At the end of each treatment period, 20 μl of MTT stock solution (5 mg/ml) was added to well and again left under incubation for 5 h (Varshosaz et al. 2013). Following this, all media was aspirated out and 200 μl of fresh DMSO was added to each well. The reading was taken at 550 nm (absorbance) against an appropriate blank.
Data analysis
The relationships between responses and formulation variables of all model formulations were treated by Design-Expert® software. Statistical analyses, including stepwise linear regression and response surface analysis, were conducted. The significant terms were chosen for the final equations. Quadratic models were found suitable for the analysis. The best fitting mathematical model was selected based on the comparisons of several statistical parameters, including the coefficient of variation (c.v.), the multiple correlation coefficient (R2), and the adjusted multiple correlation coefficient (adjusted R2), proved by Design-Expert® software (Version 9). The significance of differences was evaluated using the Student's t-test and one way ANOVA, at the probability level of 0.05.
Results
Preparation and characterization of Nano-Qu
Several methods have been reported for nanoencapsulation of drugs into polymeric matrices, such as emulsification, interfacial deposition, or nanoprecipitation. A modified version of the nanoprecipitation method was adopted for the synthesis of Qu-NP at room temperature. PLGA and Qu were mixed thoroughly in the solvent (i.e., acetone), and the mixture was then injected under pressure into the water. A volume of water was added to get a homogeneous nanocolloidal suspension. The morphology and size of the prepared PLGA NPs encapsulating Qu were assessed by TEM and DLS. Qu-loaded PLGA NPs revealed a mean diameter in the range of 100–110 nm, and a low polydispersity index (PDI) (< 0.1), that suggest that NPs are homogeneous in nature. Statistically, no significant difference was observed on the average diameter of the FA-conjugated NPs (100 nm) and that of the MA-conjugated NPs (103 nm). Conjugation of MA and FA to the PLGA NPs did not cause a change in the PDI of the NPs (0.06) when compared to plain Qu-PLGA NPs (0.065). Qu-loaded PLGA NPs have an anionic surface charge of − 20.98 ± 2.2 mV. The physical adsorption (PA) of MA leads to significant increase in the zeta potential of MA-Qu-PLGA (− 28.43 ± 1.65 mV), while a rise in the zeta potential was also noticed by conjugating FA-Qu-PLGA (− 23.20 ± 3.5 mV). Qu was found to be only sparingly soluble in water, and hence the choice was restricted to organic solvents. The important factor here is that the nanoprecipitation method renders a surfactant-free preparation of NPs ().
Bioconjugation with folate/mannose: conjugation chemistry
In order to achieve the targeting ability, QU-PLGA NPs were conjugated with FA and MA using nanoprecipitation techniques, with slight modification. Two different approaches were employed for ligand conjugation, and the prepared NPs were then evaluated through their physiochemical characterization and in vitro studies. A system of drug delivery through NPs implies that a systematic characterization is needed to confirm the characteristic properties of the NPs. Size is the most important parameter in controlling the release profile, degradation rate, cellular uptake, and biodistribution. The zeta potential is also a crucial factor to be considered in the characterization of NPs. FTIR was done to confirm the PA of mannose to the Qu-NP, and the conjugation of FA to the PLGA NPs through the crosslinking of EDC-NHS on the surface of the NP.
Structural confirmation: TEM analysis, FTIR analysis, & XRD dtudy
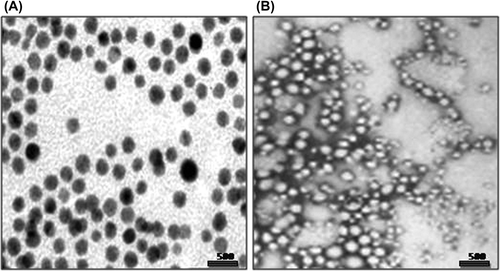
TEM by negative staining confirmed the actual size and shape of ligand-conjugated Qu-loaded NPs.
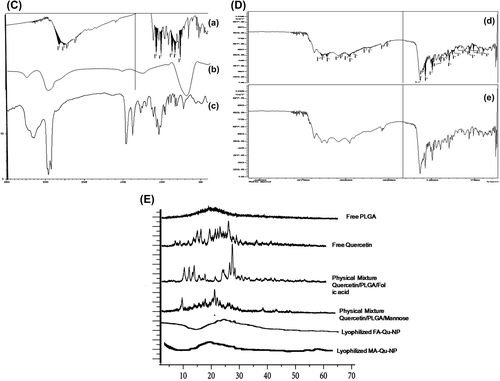
FTIR spectroscopy was used to identify the infrared absorption peaks of Qu, PLGA, FA, MA, FA-Qu-PLGA, and MA-Qu-PLGA NPs, to confirm the conjugation between the Qu-NP and the FA as well as the MA ligand. The spectral scan was carried out in the frequency ranging from 4000 to 400 cm− 1, using a Shimadzu FTIR-1700. The FTIR data showed that there was a conjugation in the case of FA-Qu-PLGA NPs, which was confirmed by the characteristic peaks of PLGA, Qu, and FA. The FTIR analysis showed characteristic bands of –CH–CH2–CH3 at 2850–3000 cm− 1 of PLGA.
Similarly, the characteristic –OH group stretching for Qu was observed at the range of 3700–300 cm− 1. Another important peak significant for the distribution of the aromatic ring was observed at 1200–900 cm− 1. Qu-loaded PLGA NPs showed band stretching for the C = O group at a range of 1700–1800 cm− 1. Folate showed a significant peak at 3545 cm− 1, corresponding to the presence of the hydroxyl group (O–H). MA showed a characteristic peak at 1600 cm− 1. The conjugation between MA and PLGA was confirmed by the availability of a sharp peak between 1625 and 1630 cm− 1, shown in .
Figure 2. TEM Images of (A) FA-Qu-PLGA NP, (B) MA-Qu-PLGA NP, (C) & (D) FTIR Spectra of Qu, MA, FA, and their conjugates, (E) XRD Spectra of free PLGA, physical mixture of Qu/PLGA/FA, and Qu/PLGA/MA, and their lyophilized NPs.
The XRD study revealed that the patterns for natural flavonoidal substances, that is, Qu, physical mixtures of Qu with PLGA, free PLGA NPs, free folic acid, free MA, FA-conjugated Qu-loaded PLGA NPs, and MA-conjugated Qu-loaded PLGA NPs, are shown in . The absence of peaks in the XRD pattern for PLGA confirmed its amorphous nature, while the natural flavonoids Qu, FA, and MA showed characteristic peaks confirming their crystalline structures. The physical mixtures of Qu, FA, and PLGA, and Qu, MA, and PLGA, also showed different signals, with variance from the original peaks and the emergence of newer peaks. These results establish the results obtained from the FTIR study, which implied the existence of weaker interactions between Qu, folic acid, and MA, with the PLGA. The nonappearance of signals related to the Qu as a natural flavonoid confirms that the conjugated NPs exhibit an amorphous nature inside the polymer matrix.
Optimization of the formula
The central composite design of the response surface methodology provides an alternative method to evaluate the possible effects of a number of variables with a limited number of experiments. The process of optimization was carried out in two sets. For the optimization of FA-conjugated Qu-NPs, four independent variables were selected, which can influence the formation of NPs (). These were the drug/polymer ratio (A), sonication time (B), volume of conjugating agents (C), and concentration of ligand (D) (). For the MA-attached Qu-NPs, three independent variables were again selected, like the drug/polymer ratio, sonication time, and concentration of ligand used (). Their effect on the response variables were studied. The distinctive response variables were particle size, zeta potential, EE, and amount of FA/MA conjugation. The observed values, along with the predicted values, are depicted in , and experimental factors in coded and actual levels are shown in , below.
Figure 3. (A) Three-dimensional (3D) reponse surface plots showing the effect of the variable on the response (Zeta Potential-FA-Qu-PLGA) nanoparticle. (B) Three-dimensional (3D) response surface plots showing the effect of the variable on the response (Zeta Potential-Ma- Qu-PLGA).
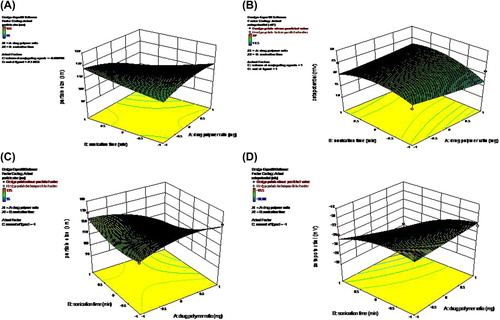
Table I. Summarizes the physiochemical characteristics of the prepared nanoformulation, before and after incorporation of FA and MA.
Table II. Solubility profile of quercetin (Qu) and its nanoformulations in different solvents.
Table III. Independent variables and their corresponding levels of Qu-PLGA NP preparation for FA binding.
Table IV. Independent variables and their corresponding levels of Qu-PLGA NP preparation for MA binding.
Table V. Predicted optimum ranges of the independent variables (MA-Qu-PLGA).
Table VI. Predicted optimum ranges of the independent variables (FA-Qu-PLGA).
Amount of conjugation
The amount of FA conjugation was found to be 11.46 ± 2.28 μM/mg, while that of MA attachment was found to be 20.69 ± 3.7 μM/mg. The particle size remained in the nano range, even after conjugation (MA-Qu-PLGA 102.39 ± 7.64, and FA-Qu-PLGA 100.3 ± 7.26). Through 3D response surface graphs, it was observed that the higher the drug concentration, the lower the EE. Similarly, higher drug/polymer ratio resulted in higher EE, and also an increase in particle size. There is a significant effect of the sonication time on the size of NPs. The value of PDI was found to be 0.06, indicating the narrow zone of size distribution. The zeta potential for MA-Qu-PLGA was found to be − 28.43 mV, and that of FA-Qu-PLGA was 23.20 ± 3.5 mV, which is lower than − 30 mV, showing good physical stability. Similarly the particle size for the MA-Qu-PLGA NP was found to be 102.39 nm, and that of FA-Qu-PLGA was found to be 100.3 nm, indicating that particles are in the nano range after conjugation ().
There was a slight difference between the EE values of the ligand-conjugated NPs (). Although there was a remarkable difference in the amount of ligands attached or conjugated to the NPs, the variables of optimization showed a good model fit. The ANOVA for response surface quadratic model was employed. The lack of fit was not significant at 95% confidence level. All other parameters were significant, at p < 0.05. The following polynomial equation was generated by the statistical analysis.
In vitro release
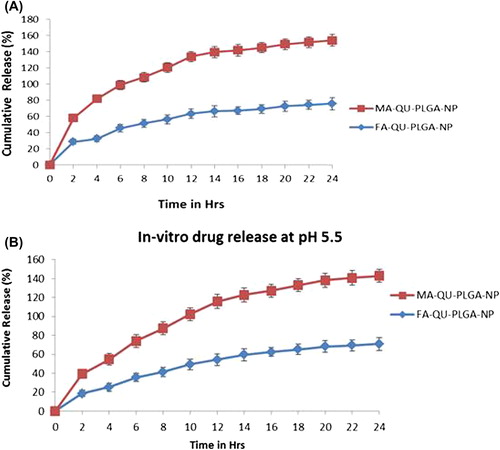
The in vitro drug release profile of nano Qu was investigated in PBS at 37°C. The cumulative percentage release is shown in . The study was carried out at the physiological and acidic pH of 5.5 and 5.6 respectively, by dispersing lyophilized NPs into PBS contained in microcentrifuge tubes. The drug release profile measured in both pH conditions followed similar kinetics, with a cumulative release of ∼70% of initial drug loading at the end of 24 h. As evident from the figure, the initial drug release amounted to ∼30% at a pH of 5.6 (), and ∼20% at a pH of 5.5, for FA-Qu-PLGA NPs, while it remained about ∼32% for MA-Qu-PLGA NPs at a pH of 5.6, and nearly ∼25% at a pH of 5.5 in 24 h. A slower and approximately steady state (lag state) with sustained release kinetics was observed during the later stages (). The initial release could be due to the drug diffusion through the superficial layers of the polymeric particles, whereas the drug release from the inner polymer matrix leads to a cumulative sustained release profile. The cumulative release of Qu from FA-conjugated PLGA NPs was measured to be 77.87 ± 1.85%, and 69.23 ± 1.54%, for pH 5.6 and 5.5 respectively, whereas for MA-attached PLGA NPs, the release was found to be 79.23 ± 1.32%, and 72.3 ± 1.72%, for pH 5.6 and 5.5 respectively, at the end of 24 h. It was noticed that the drug release was lower when the pH of the release medium was acidic.
MTT assay
Similarly, HaCaT cells and A431 cells were also incubated with different concentrations of FA-Qu-NP, MA-Qu-NP, Qu-NP, and free Qu for 48 h, and then examined by measuring absorbance. A reduction in OD550 NM was observed for the MTT assay. The results showed that in both the cell lines, FA-Qu-NP and MA-Qu-NP exhibited a significant dose-dependent activity, in comparison to the Qu-NP and free Qu.
Discussion
In the progress of UV radiation-induced skin carcinoma, the macrophage migration factor (MIF) plays a crucial role. MIF is a lymphokine that is a potent activator of macrophages in vivo, and is expressed in the skin as well. It is found to be involved in various dermatologic disorders too. The choice of MA as a ligand is based on the key findings that MA receptors are highly expressed on macrophages, becoming hot targets for carbohydrate-based delivery systems, as they internalize the MA through receptor-mediated endocytosis. Similarly, FA receptors are also expressed by the activated macrophages and are known to have higher affinity towards folic acid, thus allowing easy detection and targeting of cancerous cells.
In our study, we aimed to utilize both these approaches to design and develop a system of targeted drug delivery of natural bioactives for skin cancer. The first and foremost problem was to increase the solubility and stability of Qu. Qu, being a natural bioactive, has a wide spectrum of activity, from being antioxidant to anticancer in nature. Its potential is limited, due to its poor aqueous solubility and low instability in the physiologic medium, leading to low bioavailability and rapid metabolism. To overcome these problems, several methods were developed in the course of time, with the objective of targeting. In this regard, Varshosaz et al., in 2013, formulated solid lipid NPs of Qu, for treating hepatocellular carcinoma. They compared the effect of three different sterols, namely cholesterol, stigmasterol, and stigmastanol, in enhancing the penetration of Qu inside the cell.
In our study, we encapsulated Qu in the form of a polymeric NP (PLGA-Qu). This encapsulation increased its solubility and stability in the aqueous as well as in the physiologic medium. The relationship of solubility with the zeta potential for MA-Qu-PLGA was found to be − 28.43 mV, and that of FA-Qu-PLGA 23.20 ± 3.5 mV, which is lower than − 30 mV, showing good physical stability. The second aspect was to target these NPs. In a study, Sreeja et al., in 2010, applied the FA conjugational chemistry for targeting grape seed extract (GSE), and evaluated its anticancer potential in LKB, MCF7, and A549 cell lines. They claimed the successful targeting of GSE, employing EDC-NHS conjugational chemistry. In another study by Nahar et al., MA-anchored polymeric NPs of amphotericin were developed, to target macrophages. They employed a direct complexation technique to anchor MA to the polymeric NP, to treat leishmaniasis. All these studies direct our attention towards the fact that targeting of natural bioactives can be done for the purpose of treating skin cancer by considering the above approach. To achieve this, FA was biosynthesized separately, and MA was anchored to the NP by a modified PA technique. Here, the controversial factor was the particle size. The size of the particles was restricted to a narrow range because a low nanosize (less than ∼90 nm) would facilitate their entry into systemic circulation, while our concern relates to its retention on the skin's surface, epidermal, and dermal regions. The amount of FA conjugation was found to be 11.46 ± 2.28 μM/mg, while that of MA attachment was found to be 20.69 ± 3.7 μM/mg. The particle size remained in the nano range, even after conjugation (MA-Qu-PLGA 102.39 ± 7.64, and FA-Qu-PLGA 100.3 ± 7.26). The cumulative release of Qu from FA-conjugated PLGA NPs was measured to be 77.87 ± 1.85% and 69.23 ± 1.54%, for pH levels of 5.6 and 5.5 respectively, whereas for MA-attached PLGA-NPs, the release was found to be 79.23 ± 1.32% and 72.3 ± 1.72% for pH levels of 5.6 and 5.5 respectively, at the end of 24 h. TEM and FTIR studies further confirmed the actual size, shape, and conjugation of the ligand-conjugated Qu-loaded NPs. While in the MTT assay, we found that the cells’ viability pattern was A431 > HaCaT cell lines. There may be two reasons behind the effect of MA-Qu-NP and FA-Qu-NP conjugates in vivo. Firstly, it may be due to the activation of macrophages. Various studies have shown that the FA receptor is expressed by activated macrophages. Similarly, the carbohydrate binding site is also available at different macrophages. On chronic exposure to UV radiation, this leads to enhanced MIF (macrophage migration inhibitory factor) production, which is highly expressed on epidermal keratinocytes and fibroblasts. Secondly, the cargoes were designed in such a way that Qu dually acts as an anti-MIF agent which influences its production, as well as exhibiting MIF tautomerase activity. Our study is a minor step towards designing a targeted nanosystem of natural bioactives. Although various parameters remain to be analyzed, to arrive at a conclusion about the reliability of these systems, macrophage-targeting will certainly open new horizons for drug targeting in the near future.
Conclusion
On comparing the results obtained from the application of a central composite design, between the predicted and actual responses, we can come to the conclusion that surface response methodology is helpful in achieving suitable targeting of NPs to obtain a sustained-release drug delivery system. In this study, Qu-loaded NPs were prepared by a modified nanoprecipitation technique, where no surfactant was required. Here, we came across the findings that the concentration of the polymer in the drug/polymer ratio played a crucial role in deciding the size and stability of NPs, with a significant effect on drug EE. A variation was observed in the zeta potential and PDI of the Qu NP. Similarly, there was a significant impact of sonication time and concentration of conjugating moieties on the process of attachment of ligands to the Qu-loaded NP. Two possible explanations of our findings exist. The first is that during sonication, the particle breaks down into smaller ones, providing more surface area for attachment (as in the case of MA incorporation by the PA method), and secondly, the EDC-NHS conjugational chemistry, which activates the carboxylic end of the PLGA, is more reactive in becoming conjugated with the FA.
In summary, we formulated Qu-loaded PLGA NPs, by following a very simple technique of nanoprecipitation, and further conjugated them with two different ligands, viz. FA and MA, separately. In comparison to previous studies, we found that targeting of the Qu-NPs can be achieved through these ligands, and they satisfy the desired characteristic properties intended for targeting. Although several study parameters are required to confirm the potentialities of these delivery systems, it may be possible that a targeted drug delivery system for natural bioactives could be formulated in the near future.
Acknowledgements
The authors acknowledge the University Grant Commission-SAP [F. No.3-54/2011(SAP-II)], New Delhi, India, and the University Grant Commission (UGC), New Delhi, Under MRP Scheme Major Research project, F. No 39-170/2010 (SR), for financial assistance. One of the authors extend their gratitude towards the head of the Cosmetic Lab, University Institute of Pharmacy, Pt., Ravishankar Shukla University, Raipur (C.G.), for providing facilities to carry out research work.
Declaration of interest
The authors report no declarations of interest. The authors alone are responsible for the content and writing of the paper.
References
- Soppimath KS, Aminabhavi TM, Kulkarni AR, Rudzinski WE. 2001. Biodegradable polymeric nanoparticles as drug delivery devices. J Control Release. 70:1–20.
- Makadia HK, Siegel SJ. 2011. Poly Lactic-co-Glycolic Acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers (Basel). 3:1377–1397.
- Muller RH, Petersen RD, Hommoss A, Pardeike J .2007. Nanostructured lipid carriers (NLC) in cosmetic dermal products. Adv Drug Deliv Rev. 59:522–530.
- Wissing S, Müller R. 2002. The influence of the crystallinity of lipid nanoparticles on their occlusive properties. Int J Pharm. 242:377–379.
- Schafer-Korting M, Mehnert W, Korting HC. 2007Lipid nanoparticles for improved topical application of drugs for skin diseases. Adv Drug Deliv Rev. 59:427–443.
- Uner M. 2006. Preparation, characterization and physico-chemical properties of solid lipid nanoparticles (SLN) and nanostructured lipid carriers (NLC): their benefits as colloidal drug carrier systems. Pharmazie. 61:375–386.
- Han F, Li S, Yin R, Liu H, Xu L. 2008. Effect of surfactants on the formation and characterization of a new type of colloidal drug delivery system: nanostructured lipid carriers. Colloids Surf A Physicochem Eng Asp. 315:210–216.
- Lin X, Li X, Zheng L, Yu L, Zhang Q, Liu W. 2007. Preparation and characterization of monocaprate nanostructured lipid carriers. Colloids Surf A Physicochem Eng Asp. 311:106–111.
- Lee JW, Lu JY, Low PS, Fuchs PL. 2002. Synthesis and evaluation of taxol–folic acid conjugates as targeted antineoplastics (dagger). Bioorg Med Chem. 10:2397–2414.
- Wang AZ, Gu F, Zhang L, Chan JM, Radovic-Moreno A, Shaikh MR, Farokhzad OC. 2008. Biofunctionalized targeted nanoparticles for therapeutic applications. Expert Opin Biol Ther. 8:1063–1070.
- Mittal V, Patel SJ, Sheth SK. 2010. Development and characterization of folate targeted nanoparticle drug delivery system. Intern. J Pharma Bio Sci. 1:1–12
- Stella B, Arpicco S, Peracchia MT, Desmaële D, Hoebeke J, Renoir M. 2000. Design of folic acid-conjugated nanoparticles for drug targeting. J Pharm Sci. 89:1452–1464.
- Narayanan S, Binulal NS, Mony U, Manzoor K, Nair S, Menon D. 2010. Folate targeted polymeric ‘green’ nanotherapy for cancer. Nanotechnology. 21:285107.
- Nobs L, Buchegger F, Gurny R, Allemann E. 2004. Poly(lactic acid) nanoparticles labeled with biologically active neutravidin for active targeting. Eur J Pharm Biopharm. 58:483–490.
- Nukolova NV, Oberoi HS, Cohen SM, Kabanov AV, Bronich TK. 2011. Folate-decorated nanogels for targeted therapy of ovarian cancer. Biomaterials. 32:5417–5426.
- Lu Y, Ding N, Yang C, Huang L, Liu J, Xiang G. 2012. Preparation and in vitro evaluation of a folate-linked liposomal curcumin Formulation. J Liposome Res. 22:110–119.
- Zhang L, Zhu W, Yang C, Guo H, Yu A, Ji J. 2012. A novel folate- modified self-microemulsifying drug delivery system of curcumin for colon targeting. Int J Nanomed. 7:151–162.
- Dosio F, Milla P, Cattel L. 2010. EC-145, a folate-targeted Vinca alkaloid conjugate for the potential treatment of folate receptor-expressing cancers. Curr Opin Investig Drugs. 11:1424–1433.
- Xia Q, Hao X, Lu Y, Xu W, Wei H, Ma Q, Gu N. 2008. Production of drug-loaded lipid nanoparticles based on phase behaviors of special hot microemulsions. Colloids Surf A: Physicochem Eng Asp. 313–314:27–30
- Hentschel A, Gramdorf S, Muller RH, Kurz T. 2008. Carotene-loaded nanostructured lipid carriers. J Food Sci. 73:N1–N6.
- Wu G, Zhou F, Ge L, Liu X, Kong F. 2012. Novel Mannan-PEG- PEModified bioadhesive PLGA nanoparticles for targeted gene delivery. J Nanomater. 2012:1–9.
- Teeranachaideekul V, Muller RH, Junyaprasert VB. 2007. Encapsulation of ascorbyl palmitate in nanostructured lipid carriers (NLC)–effects of formulation parameters on physicochemical stability. Int J Pharm. 340:198–206.
- Mehnert W, Mader K. 2001. Solid lipid nanoparticles: production, characterization and applications. Adv Drug Deliv Rev. 47:165–196.
- Abu-Izza KA, García-Contreras L, Lu DR. 1996. Preparation and evaluation of sustained release AZT-loaded microspheres: optimization of the release characteristics using response surface methodology. J Pharm Sci. 85:144–149.
- Kassama LS, Shi, J, Mittal GS. 2008. Optimization of supercritical fluid extraction of lycopene from tomato skin with central composite rotatable design model. Sep Purif Technol. 60:278–284.
- Huang YB, Tsai YH, Yang WC, Chang JS, Wu PC, Takayama K. 2004. Once-daily propranolol extended-release tablet dosage form: formulation design and in vitro/in vivo investigation. Eur J Pharm Biopharm. 58:607–614.
- Ahn JH, Kim YP, Lee YM, Seo EM, Lee KW, Kim HS. 2008. Optimization of microencapsulation of seed oil by response surface methodology. Food Chem. 107:98–105.
- Zhang X, Liu J, Qiao H, Liu H, Ni J, Zhang W, Shi Y. 2010. Formulation optimization of dihydroartemisinin nanostructured lipid carrier using response surface methodology. Powder Technol. 197:120–128
- Liu CH, Wu CT. 2010. Optimization of nanostructured lipid carriers for lutein delivery; Colloids Surf A Physicochem Eng Aspects. 353:149–156
- Gambhire M, Bhalekar SMR, Gambhire VM. 2011. Simvastatin loaded Solid lipid nanoparticles: Formulation optimization using Box Behnken design, characterization and in vitro evaluation. Current Pharma Res. 1:157–164.
- Ghasemian E, Vatanara A, Najafabadi AR, Rouini MR, Gilani K, Darabi M. 2013. Preparation, characterization and optimization of sildenafil citrate loaded PLGA nanoparticles by statistical factorial design. DARU J Pharm Sci. 21:68.
- Chidambaram M, Manavalan R, Kathiresan K. 2011. Nanotherapeutics to overcome conventional cancer chemotherapy limitations. J Pharm Pharm Sci. 14:67–77.
- Ungaro F, d’Angelo I, Miro A, La Rotonda MI, Quaglia F. 2012. Engineered PLGA nano- and micro-carriers for pulmonary delivery: challenges and promises. J Pharm Pharmacol. 64: 1217–1235.
- Cooper DL, Harirforoosh S. 2014. Design and optimization of PLGA-Based diclofenac loaded nanoparticles. PLoS ONE. 9:e87326.
- Yadav KS, Sawant KK. 2010. Modified nanoprecipitation method for preparation of cytarabine-loaded PLGA nanoparticles. AAPS Pharm Sci Tech. 11:1456–1465
- Ahlin P, Kristl J, Kristl A, Vrecer F. 2002. Investigation of polymeric nanoparticles as carriers of enalaprilat for oral administration. Int J Pharm. 239:113–120.
- Varshosaz J, Jafarian A, Salehi G, Zolfaghari B. 2013. Comparing different sterol containing solid lipid nanoparticles for targeted delivery of Quercetin in hepatocellular carcinoma. J Liposome Res. 22–28.