Abstract
White blood cells (WBCs) or leukocytes are an important part of the body's defense against infectious organisms and foreign substances. WBC segmentation is a challenging issue because of the morphological diversity of WBCs and the complex and uncertain background of blood smear images. The standard ELM classification techniques are used for WBC segmentation. The generalization performance of the ELM classifier has not achieved the maximum nearest accuracy of image segmentation. This paper gives a novel technique for WBC detection based on the fast relevance vector machine (Fast-RVM). Firstly, astonishingly sparse relevance vectors (RVs) are obtained while fitting the histogram by RVM. Next, the relevant required threshold value is directly sifted from these limited RVs. Finally, the entire connective WBC regions are segmented from the original image. The proposed method successfully works for WBC detection, and effectively reduces the effects brought about by illumination and staining. To achieve the maximum accuracy of the RVM classifier, we design a search for the best value of the parameters that tune its discriminant function, and upstream by looking for the best subset of features that feed the classifier. Therefore, this proposed RVM method effectively works for WBC detection, and effectively reduces the computational time and preserves the images.
Introduction
The immune system, which is the third line of defense in the human body, protects the body from viruses, bacteria, and pathogens. This natural defense identifies and eliminates abnormal cells, such as tumor cells. The immune system consists of immune organs, immune cells, and immune molecules. White blood cells (WBCs) are the principal components of immune cells, and play an important role in our body's immunity. In fact, WBCs normally have a constant concentration in the human blood. If the WBC count exceeds the normal range, then health problems may occur. The morphologic analysis of WBCs is one of the basic steps in pathologic blood analysis. Automated methods have been developed in recent years, to overcome the limitations of manual methods which can cause calculation inaccuracies. Automated devices use the principle of the light scattering method to calculate red blood cells (RBCs) and WBCs. Actually, WBCs can be divided into five types, namely, basophils, eosinophils, neutrophils, lymphocytes, and monocytes. The first three types are granular, and the last two types are non-granular. Distinguishing these types by using the light scattering method proved unsuccessful in this study. Thus, we used image analysis, which can provide human-like assessments, to overcome this problem. This method uses the high resolution and flexible extracted features of computer vision to improve work efficiency and accuracy. Researchers are currently focused on developing a system that can automatically identify WBCs by using blood images. However, accurate WBC segmentation before classification still poses challenges.
In image segmentation, feature extraction is a particular variety of dimensionality reduction. When the input information to an algorithm is too huge to be processed, then the input information will be transformed into a reduced representative set of features. The transformation of the input data into the set of features is known as feature extraction. The standard ELM classifier is used for image segmentation. However, the training speed of the ELM classifier is not much improved. For this, a fast relevance vector machine (Fast RVM) offers much better testing time, compared to standard ELM classification. The Fast RVM-based classification method is more suitable for applications that necessitate, probably, less complexity and real-time classification.
This paper is organized as follows: Section “Literature survey” summarizes the concepts and literature survey. Section “Proposed methodology” discusses the proposed method, and Section “Experimental results” provides the experiments and achieved results of the proposed and existing techniques. Finally, Section “Conclusion” presents the conclusions of the work.
Literature survey
Blood cell counting is a major field of study in biomedical engineering. In cases involving human blood cell segmentation, many methods are being studied and applied, to obtain better results. CitationTulsani (2013), present an approach for counting different blood cells during blood smear test. Chronic lymphocytic leukemia (CLL) is the most common type of blood cancer in Canadian adults. The goal of CitationMohammed et al. (2013) is to reduce the over and under segmentation error of the watershed algorithm by suppressing 1% of the local minima. The objective of CitationSaba and Rehman (2013) is to provide a comparative study between artificially trained and heuristics rule-based techniques employed for pattern recognition in the state of the art technology focused on script pattern recognition. CitationMohsenzadeh et al. (2013) propose a separable model in feature and sample domains. Adopting a Bayesian approach and using Gaussian priors, the learned model by RSFM is sparse in both sample and feature domains. The proposed algorithm is an extension of the standard RVM algorithm, which only opts for sparsity in the sample domain. CitationDorini et al. (2013) approach novel methods to segment the nucleus and cytoplasm of WBCs. To segment the nucleus, the image preprocessing with SMMT has been shown to be essential to ensure the accuracy of two well-known image segmentation techniques, namely, watershed transform and level-set methods.
Recently, the extreme learning machine (ELM) for single-hidden-layer feedforward neural networks (SLFN) has been attracting attention for its faster learning speed and better generalization performance than those of traditional gradient-based learning algorithms. In this work, a hybrid learning algorithm is proposed by CitationHan et al. (2013) to overcome the drawbacks of ELM, which uses an improved particle swarm optimization (PSO) algorithm to select the input weights and hidden biases, and the Moore–Penrose (MP) generalized inverse, to analytically determine the output weights. CitationChyzhyk et al. (2013) perform the segmentation of medical images following an Active Learning approach that allows quick interactive segmentation, minimizing the requirements for intervention of the human operator. An automatic segmentation of leukocytes can assist pharmaceutical companies to take decisions in the discovery of drugs, and encourages the development of an automated leukocyte recognition system. CitationSaraswat et al. (2013) propose a novel strategy based on a differential evolution (DE) algorithm to segment the leukocytes from the images of mice skin sections stained with H&E staining and acquired at 40 × magnifications.
Medical imaging is a relevant field of application of image processing algorithms. In particular, the analysis of WBC images has engaged researchers from the fields of medicine and computer vision alike. CitationCuevas et al. (2013) presented an algorithm for the automatic detection of WBCs embedded into complicated and cluttered smear images, which considers the complete process as a circle detection problem. The objective of Mohapatra et al. (2014) is to improve the ALL diagnostic accuracy by analyzing morphological and textural features from the blood image using image processing. In this study, Mohapatra et al. (2014) investigate the use of image morphometry and pattern recognition techniques for sub typing leukemic lymphoblasts according to the French–American–British classification. A software tool for automatic classification and segmentation of 2-D/3-D medical images is given by CitationStrzelecki et al. (2013). CitationChinnathambi et al. (2014) presented the robust segmentation algorithm that can reliably separate touching cells. CitationDaniel et al. (2013) state that medical imaging is a relevant field of application of image processing algorithms. In particular, the analysis of WBC images has engaged researchers from the fields of medicine and computer vision alike. To overcome the shortcomings in the traditional WBCs identification methods based on the color or gray images captured by light microscopy, a microscopy hyperspectral imaging system was used to analyze the blood smears. The system was developed by CitationLi et al. (2013), coupling an acousto-optic tunable filter (AOTF) adapter to a microscope, driven by an SPF Model AOTF controller, which can capture hyperspectral images from 550 nm to 1000 nm with the spectral resolution of 2–5 nm.
Proposed methodology
The accurate WBC segmentation becomes an important issue because differential counting plays a major role in the determination of diseases, based on which the treatment is followed for the patients. The WBC image segmentation is based on various techniques such as ELM, RVM, and Fast RVM.
ELM
A new learning algorithm, called the ELM for SLFNs, supervise batch learning. The output of an SLFN with hidden nodes (additive or RBF nodes) can be represented by
Where ai and bi are the learning parameters of hidden nodes and βi is the weight connecting the ith hidden node to the output node. G(ai, bi, X) is the output of the ith hidden node with respect to the input x.
The ELM algorithm, which consists of only three steps, can then be summarized as
(a) ELM Algorithm: Given a training set
Activation function g(x), and hidden node number,
| (1) | Assign random hidden nodes by randomly generating parameters (ai,bi) according to any continuous sampling distribution, i = 1,…, Ñ . | ||||
| (2) | Calculate the hidden layer output matrix H. | ||||
| (3) | Calculate the output weight β: | ||||
The universal approximation capability of ELM has been analyzed by using an incremental method, and it shows that single SLFNs with randomly generated additive or RBF nodes, with a wide range of activation functions, can universally approximate any continuous target functions in any compact subset of the Euclidean space is the sigmoidal function used as activation function in ELM.
The main drawback of using ELM is that some parameters need to be tuned manually. Its performance will be affected seriously when outliers exist in the dataset. In order to rectify these drawbacks, the Fast-RVM is proposed.
Fast-RVM
The Fast RVM is proposed for the segmentation of WBC images. Modifications in RVM have much faster testing time, compared to standard RVM-based classification. The relevance vector machine (RVM) classifier is a probabilistic extension of the linear regression model, which provides sparse solutions. RVM evaluates the decision function by using only a small number of the training examples. However, training is based on different objectives.
The Fast RVM has a functional form identical to the RVM, but provides probabilistic classification. Firstly, the astonishingly sparse Relevance Vectors (RVs) are obtained while fitting the 1D histogram by Fast RVM. Finally, the whole WBC regions are segmented from the original image. It also has the advantages such as high computational efficiency and no extra parameter setting.
The RVM model y(x;w) is the output of a linear model with parameters = (w1, …, wN)T, with application of a sigmoid function for the case of classification
Where σ(x) = 1/1 + exp(x)).
(A) Algorithm for RVM
The steps are as follows:
Step 1: Select a suitable kernel function for the data set and relevant parameters. Use this kernel function to create the design matrix Φ.
Step 2: Establish suitable convergence criteria for α and β, e.g. a threshold value for change δThresh between one iteration estimation of α and the next δ = Σi = 1 ain + 1−ain so that re-estimation will stop when δ<δThresh.
Step 3: Establish a threshold value αThresh, in which it is assumed an αi is tending to infinity upon reaching it.
Step 4: Choose starting values for α and β.
Step 5: Calculate m = m = βΣΦTt and Σ = (A+βΦTΦ)− 1.
Step 6: Update
Step 7: Prune αi and corresponding basic functions where αi>αThresh.
Step 8: Repeat steps (5) to (7) until the convergence criterion is met.
Our hyper parameter values α and β, which result from the above procedure, are those that maximize our marginal likelihood and hence they are used when making a new estimate of a target value t for a new input x′:
The variance relating to our confidence in this estimate is given by:
(b) The Fast RVM algorithm
The steps are as follows:
Step 1: Form a compact histogram from a given microscopic image.
Step 2: Use the RVM to approximate the above compact histogram and obtain all the RVs.
Step 3: Seek the threshold from the RV set to ensure it to occupy the deepest concavity.
Step 4: Use the so-obtained threshold to segment the given image.
Step 5: Perform morphologic operations to the above image in order to obtain the entire connective WBC region.
Given a training data set of input-target pairs D = {(xi, yi)li=1, RVM follows the standard probabilistic formulation and assumes that the targets are samples from the model with additive noise:
Where εi are error terms which are generally assumed to be independent identically distributed Gaussian variables with mean zeros and variance σ2. An improved algorithm has been proposed for identification and classification of WBCs in digital microscopy images, using the color image segmentation method.
Experimental results
The experiments are carried out using images that are collected from a hospital. The implementation is done by using MATLAB. For each image, the hematology expert classifies the WBCs, and these are recorded to build up the database. To evaluate the results of the techniques, the experiment is conducted on various blood cell images. The blood cell image contains RBCs, WBCs, and platelets. From these, the WBC alone are segmented, and the number of WBCs detected by various techniques is compared with that actually present in the image which is manually obtained. The ground truth for the complete dataset is collected from the pathologist.
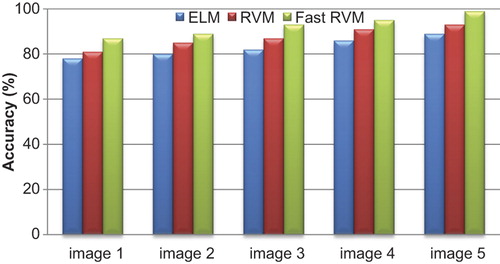
describes the accuracy values of ELM, RVM and Fast RVM classifiers for WBC image segmentation. The accuracy of the proposed method of Fast RVM is higher than the other classification techniques.
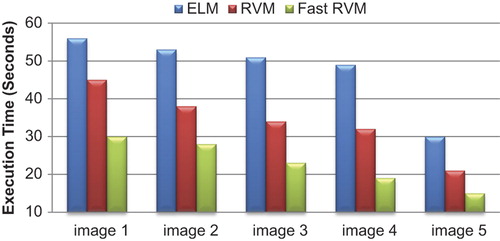
describes the execution time values of ELM, RVM, and Fast RVM classifiers for WBC image segmentation. The values for processing time are less in Fast RVM when compared to other methods like ELM and standard RVM.
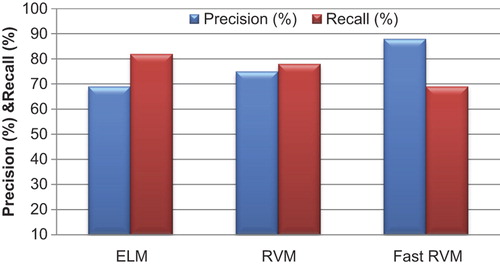
The precision, recall, and F measures are used in this research to classify image segmentation. The formulas for calculating the precision, recall, and F measures are described.
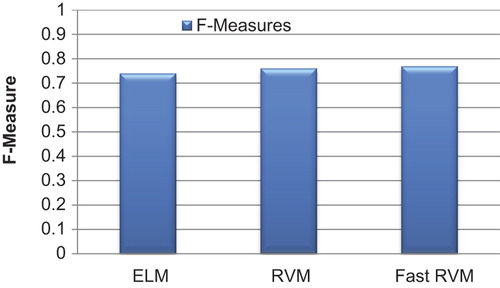
In , the precision and recall values are estimated using the formula for WBC image segmentation. The precision values are higher in the proposed Fast RVM method when compared to those obtained with other techniques, and recall values are very low in the proposed technique. In , the values for F-Measure indicate the overall higher performances of Fast RVM technique in WBC image segmentation.
The planned method uses discriminative features, which evidently contain information for better discrimination of blood cells. Further, feature selection methods, based on mutual information distribution and recursive feature removal along with Fast RVM, are used for effective classification.
Conclusion
The WBCs are segmented by using the improved algorithm. In this work, the fast RVM is proposed for WBC image segmentation. The proposed method is simple and has a fine level of accuracy, with an incredible reduction in time and complexity. With this proposed technique, the image will detect the majority of WBCs, and each detected cell is nearly complete. It has strong adaptability and high computational efficiency. Moreover, no extra parameter setting is needed. The experimental results confirm that the fast RVM classification system substantially boosts the generalization capability achievable with the ELM classifier, and it is robust against the problem of limited training availability, which may characterize pathologies of rare occurrence. It can also be seen that the Fast RVM accomplishes better and more balanced classification for individual categories as well, with a very low training time compared to ELM. The results obtained encourage future work, which may include this effective algorithm as an initial process to classify the WBCs from RBCs and platelets and hence to detect immature WBC cells. It could be improved further by better pre-processing methods and feature sets, which will be taken up in our future work.
Declaration of interest
The author reports no declarations of interest. The author alone is responsible for the content and writing of the paper.
References
- Chinnathambi K, Ramasamy A, Ramachandran P. 2014. Robust segmentation of cancer affected white blood cells using modified level set algorithm. Int J Simul Syst Sci Technol. 14:9.
- Chyzhyk D, Ayerdi B, Maiora J. 2013. Active learning with bootstrapped dendritic classifier applied to medical image segmentation. Pattern Recogn Lett. 34:1602–1608.
- Cuevas E, Oliva D, Díaz M, Zaldivar D, Pérez-Cisneros M, Pajares G. 2013. White blood cell segmentation by circle detection using electromagnetism-like optimization. Comput Math Methods Med. 2013:395071.
- Daniel Z, Oliva D, Cuevas E, Pajares G, Pérez-Cisneros M, Diaz M. 2013. White blood cell segmentation by circle detection using electromagnetism-like optimization. Comput Math Methods Med. 2013:395071.
- Dorini LB, Minetto R, Leite NJ. 2013. Semiautomatic white blood cell segmentation based on multiscale analysis. IEEE J Biomed Health Inform. 17:250–256.
- Han F, Yao H.-F, Ling Q.-H. 2013. An improved evolutionary extreme learning machine based on particle swarm optimization. Neurocomputing. 116:87–93.
- Li Q, Wang Y, Liu H, Wang J, Guo F. 2013. A combined spatial-spectral method for automated white blood cells segmentation. Opt Laser Technol. 54:225–231.
- Mohammed EA, Mohamed MMA, Naugler C, Far BH. 2013. Chronic lymphocytic leukemia cell segmentation from microscopic blood images using watershed algorithm and optimal thresholding. In Electrical and Computer Engineering (CCECE), 2013 26th Annual IEEE Canadian Conference on, pp. 1–5. IEEE.
- Mohapatra S, Patra D, Satpathy S. 2014a. An ensemble classifier system for early diagnosis of acute lymphoblastic leukemia in blood microscopic images. Neural Comput Appl. 24:1887–1904.
- Mohapatra S, Patra D, Satpathy S, Jena RK, Sethy S. 2014b. Automated morphometric classification of acute lymphoblastic leukaemia in blood microscopic images using an ensemble of classifiers. Comput Methods Biomech Biomed Eng Imaging Vis. 1–14.
- Mohsenzadeh Y, Sheikhzadeh H, Reza AM, Bathaee N, Kalayeh MM. 2013. The Relevance Sample-Feature Machine: A Sparse Bayesian Learning Approach to Joint Feature-Sample Selection. 1–1.
- Saba T, Rehman A. 2013. Effects of artificially intelligent tools on pattern recognition. IJMLC. 4:155–162.
- Saraswat M, Arya KV, Sharma H. 2013. Leukocyte segmentation in tissue images using differential evolution algorithm. Swarm Evol Comput. 11:46–54.
- Strzelecki M, Szczypinski P, Materka A, Klepaczko A. 2013. A software tool for automatic classification and segmentation of 2D/3D medical images. Nucl Instr Meth Phys Res A. 702:137–140.
- Tulsani H. 2013. Segmentation using morphological watershed transformation for counting blood cells. IJCAIT. 2:28–36.