Abstract
Omics technologies emerged as complementary strategies to genomics in the attempt to understand human illnesses. In general, proteomics technologies emerged earlier than those of metabolomics for major depressive disorder (MDD) research, but both are driven by the identification of proteins and/or metabolites that can delineate a comprehensive characterization of MDD's molecular mechanisms, as well as lead to the identification of biomarker candidates of all types—prognosis, diagnosis, treatment, and patient stratification. Also, one can explore protein and metabolite interactomes in order to pinpoint additional molecules associated with the disease that had not been picked up initially. Here, results and methodological aspects of MDD research using proteomics, metabolomics, and protein interactomics are reviewed, focusing on human samples.
Las tecnologías “ómicas” aparecieron como estrategías complementarías a la genómica en el intento de comprender las enfermedades humanas. Aunque en general las tecnologías proteómicas surgieron antes que las metabolómicas en la investigación del trastorno depresivo mayor (TDM), ambas están orientadas a la identificación de proteínas ylo metabolítos que permita esbozar una completa caracterización de los mecanismos moleculares del TDM, así como también llevar a la identificación de biomarcadores candidatos de todos los tipos: pronóstico, diagnóstico, tratamiento y estratificación de pacientes. También se pueden explorar interactomas de proteínas y metabolitos para precisar moléculas adicionales, asociadas con la enfermedad, que no han sido identificadas inicialmente. En este artículo se revisan los resultados y aspectos metodológicos de la investigación en el TDM que emplea proteómica, metabolómica e interactómica de proteínas, centrándose en muestras de humanos.
Les technologies dites « omiques » sont apparues pour compléter la génomique afin de comprendre les maladies humaines. D'une façon générale, les technologies protéomiques sont antérieures aux métabolomiques dans la recherche sur l'épisode dépressif caractérisé (EDC) mais les deux sont fondées sur l'identification des protéines etlou des métabolites pour définir une description complète des mécanismes moléculaires de l'EDC et identifier des biomarqueurs candidats de tout type, pronostique, diagnostique, thérapeutique, et pour la stratification des patients. Il est également possible d'analyser les interactomes des protéines et des métabolites afin de repérer d'autres molécules associées à la maladie et non détectées au début. Cet article étudie chez l'homme les résultats et les aspects méthodologiques de la recherche sur l'EDC par la protéomique, la métabolomique et l'interactomique des protéines.
Depression
Depression—unipolar depression, clinical depression, or major depressive disorder (MDD)—is a severe neuropsychiatric disorder that affects 350 million diagnosed patients and their families worldwide. The National Institutes of Health (NIII) estimates that 60% of people who commit suicide have MDD or another mood disorder in the USA. Additionally, the World Health Organization (WHO) predicts that by 2030, MDD will be the leading cause of global disability.Citation1 Most alarming is the fact that the main strategy of MDD management, which is antidepressant medication, shows only modest efficacy: 40% of patients do not respond to current treatments and often experience undesirable side effects.Citation2 Moreover, medication response is lengthy, with high rates of relapse and treatment resistance.Citation3
MDD's underlying molecular mechanisms are still to be unraveled. Novel technologies such as omics-based platforms may offer new insights into the pathobiologic core of MDD, as well as the possibility of discovering potential diagnostic, therapeutic, and disease course biomarker candidates.Citation4
Proteomics
Proteomics is the science that emerged from the term “proteome,”Citation5 which can be defined as the set of expressed protein by a cell, tissue, or organism, in a given moment, under a determined condition. Nowadays proteomics approaches much more than the study of the proteome, including the characterization and identification of post-translational modifications, protein-protein interaction, protein turnovers, and more.
Methodologies for proteome investigations
The identification, and eventually the quantification, of a given proteome of interest is the most popular tool in the proteomics toolbox. Two-dimensional gel electrophoresis (2DE) combined with mass spectrometry (MS) had been the basis of proteomics since its beginning. Recently, the combination 2DE-MS has been replaced gradually by shotgun proteomics (or shotgunmass spectrometry [shotgun-MS]). Both approaches have advantages and disadvantages, and their combination seems to be the best strategy.
Two-dimensional gel electrophoresis combined with mass spectrometry
The principle of 2DE, a methodology developed back in the 1970sCitation6 and further optimized since then,Citation7-Citation10 is to separate the proteins by two of their physicochemical characteristics. First, using isoelectrofocusing (IEF), proteins are separated according to their isoelectric point (pI) in a gel with an immobilized pH gradient. These proteins are washed in sodium dodecyl sulfate (SDS) solution and then separated according to their apparent molecular weight using SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins may be stained after electrophoresis, or even labeled with fluorescent dyes prior to electrophoresis, also known as 2D fluorescence difference gel electrophoresis (2D-DIGE).Citation11 Each sample has a proteome defined in a gel. Each gel is filled with dots, technically called spots, which can be compared across different gels according to their density, calculated with the help of computational software according to their intensity and volume. Spots of interest can be excised from the gels, digested, and identified by MS.
In the 1990s, the 2DE-MS approach was an attractive technique to separate thousands of proteins using relatively low amounts of samples. Nowadays, shotgun-MS techniques, which started to emerge at the end of the 1990s,Citation12 require two orders of magnitude less of samples, and the material is handled in a more automated manner. Moreover, some of 2DE's drawbacks, such as the potential overlap of proteins in a single spot, as well as the resolution of low abundant, hydrophobic, very acidic, very basic, very small, and very large proteins, can be avoided by shotgun-MS.
Shotgun-MS
In shotgun-MS approaches, gels are not needed to separate the proteins prior to their identification. The basic principle is to digest the whole proteome of interest and inject in a system that possesses high-performance liquid chromatography (HPLC) coupled online to a mass spectrometer (LC-MS). Data from shotgun-MS, which consists of chromatograms attached to mass spectra, are analyzed by complex computational algorithms to reconstruct the protein sequences based in the masses of all peptides measured and fragmented. This process is known as “bottom-up proteomics.” MS-based proteomics have rapidly developed in the past 10 years. Nowadays, a single LC-MS experiment is able to reveal 3000 to 7000 proteins in an hour, which would only be doable—if at all—by combining 2DE-MS over some weeks of work.
For proteome quantitation, there are several alternatives that can be taken into considerationCitation13 for a given LC-MS experiment, such as stable isotope labeling in vitro (ie, isotope-coded affinity tags [ICAT]Citation14 and isobaric tags for relative and absolute quantitation [iTRAQ]Citation15) or even in vivo (ie, stable isotope labeling by/with amino acids in cell culture [SILAC]Citation16 or stable isotope labeling in mammals [SILAM]), as well as diverse label-free approaches.Citation17
Shotgun-MS still presents difficulty in representing hydrophobic and low abundant proteins depending on the type of sample preparation and MS acquisition. Moreover, information of intact proteins is lost by conventional bottom-up proteomics, which can be represented by 2DE, as well as the characterization of certain protein post-translational modifications.
Proteome findings in patients with depression
Brain tissue and cerebrospinal fluid
Surprisingly, and unlike other psychiatric disorders such as schizophrenia,Citation18,Citation19 only one research group focused their efforts on the large-scale proteome investigation of postmortem human brains from depressed patients, through two articles. Samples from the dorsolateral prefrontal cortex (DLPFC) of 24 patients with MDD were compared with 12 controls using a shotgun label-free approach. Some of the protein candidates were further validated by selected reaction monitoring (SRM). Several biological functions were associated with MDD, such as energy metabolism, cellular transport, and cell communication and signaling.Citation20 Energy metabolism has already been described for a long time as a pattern for psychiatric disorders in general, via several different techniques.Citation21-Citation23 However, it has been possible to delineate exactly which energy metabolism pathways are more involved in each disorder, by using proteomics. Glycolysis is the main affected pathway in schizophrenia brains,Citation24 whereas in MDD, oxidative phosphorylation is the most affected. Not only have several subunits of oxidative phosphorylation complexes been shown to be expressed differentially, but adenosine triphosphate (ATP) levels were also determined to be lower in MDD.Citation20 Additionally, a proteomic study of a preclinical model for anxiety has shown both pathways to be differentially regulated.Citation25
Of the 24 MDD samples analyzed in the proteome study cited above, 12 presented with psychosis as onet of their symptoms (MDD-P), in contrast to the remaining 12 (MDD-NP). Interestingly, when the factor “psychosis” was taken into consideration, the proteome differences between MDD-P vs controls, or vs MDD-NP revealed similarities in proteins differentially expressed in schizophrenia, including glycolysis enzymes, which were not present when all MDD patients were compared with controls.Citation20
In the MDD brain, the histidine triad nucleotidebinding protein 1 (HINT1) was found to be increased, as confirmed by SRM-MS. On the other hand, HINT1 levels were found to be decreased in schizophrenia in the DLPFC.Citation26 It is important to note that when MDD patients were compared separately according to their psychotic symptoms, HINT1 was only observed as increased in MDD-P subjects. This protein has been associated with antidepressant- and anxiolytic-like effects,Citation27 and is thought to play a role in postsynaptic dopamine transmission. Additionally, HINT1 has been hypothesized to interfere with hypothalamic-pituitaryadrenal axis function.Citation27-Citation28
A second large-scale MS-based investigation of MDD brains was a phosphoproteome analysis also in the DLPFC. Ninety out of 802 proteins presented differential levels of phosphorylation in MDD compared with controls. The great majority of these proteins were associated with synaptic transmission, such as two subunits of clathrin and two subunits of spectrin, synapsin, and dynamin, in addition to proteins such as actin, actinin, and internexin, which are associated with cellular architecture.Citation29 These results align with the MDD proteomic study, which also shows a dysregulation of synaptic-related proteins,Citation20 especially those associated with soluble NSF attachment receptor (SNARE) function, such as synaptosomal-associated protein 25 (SNAP25), γ-aminobutyric acid receptor-associated protein-like 2 (GABARAPL2), and syntaxin 1B (STX1B). Synapsin I (SYN1), which also plays a role in SNARE function, has been found to be differentially phosphorylated in MDD brains.
Another two reports from other research groups present proteome investigations of MDD brain, but these studies focused on the analyses of schizophrenic brains, using MDD and bipolar disorder samples as controls for specificity The proteomes of the frontal cortex (FC)Citation30 and anterior cingulate cortex (ACC)Citation31 from depressed patients have been subjected to 2DE-based proteomic analyses, revealing an altered expression of dihydropyrimidinase-like 2 (DPYSL2). DPYSL2, also known as collapsin response mediator protein 2 (CRMP2), plays a range of roles, including participation in the development of the central nervous system by regulating axonal guidance, neuronal growth cone collapse, and cell migration.Citation32 Additionally, energy metabolism-related proteins such as carbonic anhydrase (CA2) and aldolase C (ALDOC) were found to be altered in both brain regions.
A study on schizophrenia biomarkers analyzed the cerebrospinal fluid (CSF) of 16 MDD patients.Citation33 These were classified as psychotic and nonpsychotic, and analyzed separately in order to observe whether the biomarkers identified for schizophrenia were specific, or if they were associated with acute psychosis. The authors concluded that the proteomic signature was specific enough to identify schizophrenia, but the number of psychotic MDD cases was too small.
Using the traditional proteomic combination of 2DEMS and shotgun-MS, the proteomes of the CSF from 12 MDD patients and 12 controls were compared in quantity and phosphorylation levels.Citation34 Eleven proteins were found to be differentially expressed by 2DE, and additionally by shotgun-MS.Citation25 Proteins were involved in neuroprotection, neurodevelopment, and sleep regulation. A particular set of proteins involved in energy metabolism—anti-pigment epithelium derived factor (PEDF), apolipoprotein E (ApoE), prostaglandin D2 synthase (PGDS), and cystatin C—were chosen to be validated byWestern blot due to the association of MDD with metabolic syndrome. Interestingly, PGDS, which was found to be downregulated in this study, was observed to be upregulated in the CSF of schizophrenia patients.Citation35 Differences in phosphorylation levels were observed for 16 proteins, some of which also had altered expression.
Blood
Unlike with preclinical models,Citation36-Citation37 few efforts have been so far invested in identifying proteomic differences in the blood of MDD patients in comparison with healthy subjects. The blood plasma proteome from 21 first-onset drug-naive MDD patients was compared with the same number of controls, employing a shotgun proteomics platform combined with iTRAQ in a hypothesis-free manner.Citation38 Further validations of protein candidates were performed by Western blot and enzyme-linked immunoadsorbent assay (ELISA).The modest number of 9 proteins were found to be differentially expressed in MDD patients, being mostly involved in lipid metabolism and the immune system, which are postulated to be involved in the early stages of MDD pathophysiology.Citation38 The importance of this study is not only to reveal potential biomarker candidates,Citation39 but also in the comprehension of MDD as a systemic disorder.
Mononuclear cells
There is a need to improve the understanding of the molecular mechanisms triggered by successful antidepressant treatment. With this in mind, a mass spectrometry-based proteome analysis of blood mononuclear cells (MNC) collected from inpatients upon admission (T0) and after 6 weeks of psychopharmacological treatment (T6) was performed. Patients were classified as good or poor responders, and their proteomic profiles were compared at T0.
Proteins related to integrin and Ras signaling exhibited different MNC expression levels at T0. In addition, a longitudinal proteomic profiling analysis (T0-T6) to investigate the biology involved in the antidepressant treatment response showed that the biological processes for good and poor responders were similar, but they presented different patterns of regulation. These results go along with the implementation of new strategies for a personalized medicine approaches by predicting which patients might respond to a specific antidepressant.Citation40
Metabolomics
One of the first reports mentioning the term “metabolomics” came out in the literature around the year 2000.Citation41-Citation42 With a similar proposal to proteomics, this technology emerged as a means of understanding biological systems and diseases in a large-scale manner, through the identification of metabolic substrates and products of a given biochemical system. The technique of metabolomics may also be of use in the research of xenobiotics, drugs, and medications.
Considering the metabolome as the metabolic state of a given physiologic status of a given cell, tissue, or organism, metabolomics is not only a complementary tool for understanding proteomics data, but also a discipline that stands on its own, able to reveal biochemical pathways involved in biological mechanisms of interest, as well as potential biomarkers.Citation43
Methodologies
Sample preparation
The sample preparation for metabolome analyses is the most important part of the study. It depends on the classes of metabolites that one wants to study, for example, general metabolome analysesCitation44 or more target-oriented analyses according to the interest of the study in detecting hydrophilicCitation45 or hydrophobicCitation46 molecules. Also, the sample preparation will depend on the type of platform to be employed, which will be discussed ahead. The most important issue during sample preparation, especially when a comparative study is performed, is to assure that samples are collected using a standardized procedure, in order to capture the same metabolomic snapshot across all samples to be analyzed. Considering the very dynamic nature of the products of metabolism, the metabolome status can change significantly and rapidly when confronted with any mild environmental stimulus; this in itself can actually be an interesting aspect to be explored, ie, metabolites' turnover rates.Citation47
Nuclear magnetic resonance spectroscopy-based metabolomics
Nuclear magnetic resonance spectroscopy (NMR) consists of the absorption and re-emission of electromagnetic radiation by atomic nuclei in a magnetic field. Molecules, here treated as metabolites, may have their metabolic fingerprint determined by this process, leading to their identification and possibly to their quantification, in a large-scale, nontargeted, and nondestructive manner.Citation48 NMR is applicable to the analyses of biofluids, cell extracts, and cell cultures, and requires almost no sample preparation.Citation49 The standard approach for metabolomic analysis using patient's samples is using proton NMR (III NMR), although other nuclides, such as 2H, 13C, 31P, 15N, and 19F, may by employed for the generation of additional information.Citation50-Citation51
Mass spectrometry-based metabolomics
MS-based metabolomics may provide a targeted or largescale metabolome analysis.Citation52 It has become an indispensable tool in metabolome/metabonome analysisCitation53,Citation54 and is generally combined online with three types of prefractionation methods: gas chromatography (GC),Citation55 HPLC,Citation56 or capillary electrophoresis (CE).Citation57 As always in analytical chemistry, each separation method has its advantages and drawbacks: GC is highly efficient, sensitive, and reproducible, but can only be performed with volatile compounds or those that can be made volatile. HPLC separation may reach a wider range of analytes, even though its resolution is poorer. In turn, CE may present superior performance regarding separation than HPLC, but it is properly applicable to charged analytes.
The advantage of MS lies in its sensitivity and throughput.Citation58 Fingerprints of metabolites can be determined for establishing metabolome libraries, which will facilitate the identification of a given metabolite.
Metabolome findings in samples from patients with depression
Although metabolomics studies in depression are rather recent—the first report came out in 2007 studying human samples—they have become popular and even more used in human samples than proteomics. Several metabolomics studies have been performed in preclinical models of depression.Citation59-Citation62
Brain tissue and CSF
No metabolomic study has been performed in brain tissue from MDD patients thus far. There is one report with CSF analyses. A targeted metabolomic analysis was carried out in the CSF of 14 unmedicated MDD patients, 14 remitted MDD subjects, and 18 healthy controls. Tryptophan, tyrosine, purine, and related pathways were analyzed, revealing higher levels of methionine, and reduced levels of tryptophan and tyrosine in remitted patients. Additionally, the same group presented altered methionine-to-glutathione ratios, suggesting alterations in methylation and oxidative stress pathways. Unmedicated MDD subjects also showed alterations in these same metabolites, but not to a statistical level.Citation63
Blood (and urine)
Blood plasma was collected from 9 elderly MDD patients, 11 remitted patients, and 10 mentally healthy subjects. After screening over 800 metabolites by GCMS, results suggested that higher concentrations of lipid metabolites and neurotransmitters, such as dicarboxylic fatty acids, glutamate, and aspartate, are associated with MDD. Interestingly, these differences are less prominent in treated patients, who presented a metabolomic panel more similar to that of control subjects.Citation64 The panel of blood plasma metabolites associated with depression changed when a second variable came into play. While in the first study, elderly patients were considered, in this other study MDD patients with heart failure were compared with nondepressed heart failure patients. Here, GC-MS and LC-MS metabolomics platforms revealed differential concentrations of certain amino acids, such as glutamate, aspartate, and cysteine. Moreover, as in the study with elderly patients, a dysfunction of fatty acid metabolism was observed, suggesting this pathway as part of a biosignature of MDD.Citation65
Aiming to implement the metabolome of MDD patients as means of diagnosis, the plasma metabolomes of MDD patients and healthy controls were compared using NMR-based metabolomics. The analyses of 58 first-episode drug-naive depressed patients, compared with 42 controls, revealed a panel of metabolites that could distinguish these two groups in a second round of experiments, using 26 samples in a blind manner.Citation66 Similarly, the urine metabolomes of 82 first-episode drug-nai've MDD patients have been compared with 82 healthy controls by NMR-based metabolomics, revealing differences in concentration of malonate, formate, N-methylnicotinamide, m-hydroxyphenylacetate, and alanine. In a multivariate analysis, these metabolites could separate MDD from healthy controls. This same panel of metabolites was then analyzed in a second set of samples composed of 44 MDD patients and 52 healthy controls in a blind manner, achieving a similar level of group distinction.Citation67 These two studies present promising findings, especially considering their capacity to distinguish MDD groups in a blind manner. However, urine studies must be performed, keeping in mind that future applications of these results demand the establishment of standard operating procedures for sample collection, due to the large metabolic variation in urine composition.
Considering that a significant number of MDD patients do not respond to the current medications, the likelihood for a successful response has been evaluated using metabolomics. Serum metabolomes from 43 MDD patients treated with sertraline were compared before the initiation of treatment, with a group of 46 subjects receiving a placebo, using liquid chromatography electrochemical array. The metabolome profiles partially separated responders from nonresponders by employing multivariate analyses. The metabolites that contributed the most to the separation of responders from nonresponders were phenylalanine, tryptophan, purine, and tocopherol. Additionally, dihydroxyphenylacetic acid, tocopherols, and serotonin were more relevant to the separation of the medication and placebo groups.Citation68 In a more extensive study, the metabolome profiles of the serum from MDD patients treated with sertraline or placebo have been analyzed and quantified by GC-MS at three time points: prior to medication, and 1 and 4 weeks after medication. Sertraline- and placebo-induced differences in metabolites were related to tricarboxylic acid cycle (TCA), urea cycle, fatty acids and intermediates of lipid biosynthesis, amino acids, sugars, and gutderived metabolites, with more pronounced differences after 4 weeks. More specifically, sertraline showed effects on ATP-binding cassette (ABC) and solute transporters, G signaling molecules, and fatty acid metabolism. The increasing effect of the drug after 4 weeks of treatment is in line with the delayed clinical effect of the medication.Citation69
Results discussed in this topic go towards translational strategies with the potential of future clinical implementation. As for most psychiatric disorder studies, follow-up of these results is necessary.Citation70
Protein interactomics
The concept of the interactome considered here concerns the complete set of molecular interactions of a given protein in a given cell or other biological environment. Hence, all proteins, and eventually metabolites, that interact with a given protein of interest, promoting, regulating, and inhibiting its activity or expression, is part of its interactome. Establishing a protein interactome may be informative about the protein function and all molecular mechanisms in which it is involved. Additionally, the study of protein interactomes in diseases may reveal dysfunctional pathways, their regulation, and the possible role that protein partners play in the disease.Citation71
Methodologies
Protein interactome studiesCitation72 have been mainly performed employing yeast two-hybrid screening (Y2H)Citation73 and tandem affinity purification (TAP).Citation74-Citation75 Coimmunoprecipitation (coIP) has also been used as a reliable method for studying protein interactomes.Citation76-Citation77 In addition, there have been efforts invested in the establishment of algorithms that can predict the network of interactions of a given protein, including its 3D modeling, which is pivotal not only for the characterization of its roles in the cell, but also to reveal potential targets for drug discovery and design.Citation78-Citation80
Protein interactome in patients with depression
The phosphatidylinositol 3-kinase and the mammalian target of rapamycin pathway—PI3K-mTOR—plays a central role in the therapeutics of MDD through the activation of immune cells via inflammatory cytokines.Citation81 Thirty-three components of the PI3K-mTOR pathway have been targeted for a large-scale interactome analysis employing Y2H screen. More than 800 interactions to the PBK-mTOR pathway have been identified, including 67 new interactions. Further validations suggest that deformed epidermal autoregulatory factor- 1 (DEAF1) is a substrate for glycogen synthase kinase-3 (GSK3) A and B, and that this protein might be a therapeutic target of lithium treatment for MDD.Citation82
A systematic network and pathway analysis of MDD candidate genes has been constructed, based on a set of genes proposed to be associated with MDD in association, linkage, and gene expression studies of humans and animals.Citation83 An overlap of MDD's molecular features with schizophrenia has been observed. Moreover, the authors proposed neurotransmission- and immune system-related pathways as the most representative biological processes involved in MDD. Even though these are processes previously shown as involved in MDD by other fields of study,Citation84 this in silico interactome study has pinpointed the role players in the dysregulation of these pathways, which is an important example of the information that omics technologies are able to provide.Citation85
Integration of omics technologies
The overall understanding of a biomolecular system can be obtained by the integration of large-scale analyses coming from different sources, such as proteomics, metabolomics, and protein interactomics. Nowadays, considering the huge datasets that can be generated by each one of these analytical chemistry platforms, several computational tools and algorithms have been developed to integrate these results. As an example, the Webbased tool STRING (Search Tool for the Retrieval of INteracting Genes/proteins)Citation86 has been used to analyze the differentially expressed proteins found in the DLPFC of MDD patients, as described earlier.Citation20 As can be seen in STRING'S algorithm proposes several interactions among these proteins. The greater the number of colored lines connecting two proteins, the stronger the suggested evidence of their interaction.
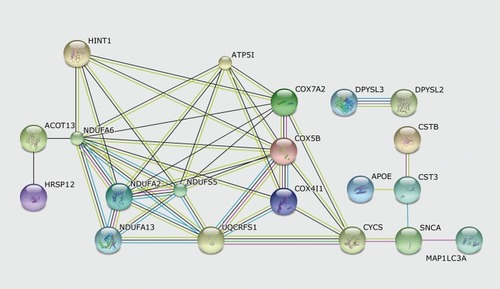
In this case, some connections are evident: for example it is known that the protein subunits of NADH dehydrogenase (ubiquinone) are all part of the complex I of the respiratory chain or the proteins COX, which are components of cytochrome c oxidase. Other interactions may be informative: for example, what is the nature of the connection between HINT1, an antidepressant-associated protein, to cytochrome c oxidase? Could this protein interfere with the mitochondrial metabolism?
It is important to note however, that the so called “interaction” may have several different levels of evidence: briefly, this interaction can be the result of an experimentally proven interaction, the two proteins may have been mentioned in a given scientific publication, or another computational algorithm may have suggested their interaction. Thus, especially in the last scenario, for which there is no experimental proof that such interaction really exists, this data must be interpreted carefully. Informative tools such as STRING, Cytoscape,Citation87 Ingenuity Pathway Analyses (Ingenuity® Systems), and Pathway Studio (Ariadne Genomics) have become popular lately, and indeed facilitate the understanding of a given molecular process. However, the final curator of these results is the researcher using it, and this step is a must. Having established a dataset of lines and connections does not mean that this represents, per se, a meaningful interactome.
Some of the tools mentioned above can also deal with drug metabolites, and even suggest interactions of proteins and metabolites of interest with known drugs. These can be informative pieces of evidence to be further investigated in the laboratory, depending on the nature of the interaction proposed by the computational tool. Here lies the beauty of the large-scale studies: generating, with parsimony, new hypotheses to be further investigated. Further information on integrative systems biology can be found in refs 88 and 89.
Conclusions and perspectives
The omics technologies applied to studies of human samples as discussed here lead to modest, but new, hypotheses. These have helped the understanding of the molecular mechanisms of MDD. As discussed above, the overall dysfunction of oxidative phosphorylation, which contrasts with the pathways noted in schizophrenia, together with the differential expression and phosphorylation of a number of synaptic proteins, may warrant further investigation regarding these particular targets. Data reviewed here must be combined with information obtained from preclinical models.Citation25,Citation37,Citation80-Citation95 These have the advantage of showing fewer confounding factors than human samples. Their limited biomechanical range must be noted, since not all features of a complex human disease such as MDD can be considered. Omics technologies, particularly metabolomics, can also be employed in the development of innovative medications, which are urgently needed.Citation96
With regards to biological markers of depression, the findings are still preliminary.Citation97 In contrast to what was expected, the identification of such biomarkers seems to be more complex than anticipated.Citation98- Citation100 An example is the recent withdrawal of VeriPsych, which was the only commercially available test biomarker for a psychiatric condition. Hie molecular overlap among psychiatric disorders makes the task of developing diagnostic tools very challenging. MDD patients who present with similar symptoms may have completely distinct biochemical signatures: some may have become depressed due to immune system-related dysfunctions, while others may have had their energy metabolism affected. Additionally, the different biological factors unrelated to the disease, such as cigarette smoking and alcohol consumption, must be taken into account carefully. Among the most wanted biomarkers are those associated with the prediction of a successful drug response. MDD treatment is lengthy, and after several weeks, about 40% of patients do not respond to current medications. The formula “one treatment suits them all” does not fit. Biomarkers to identify subgroups of patients and predict therapeutic response are needed to achieve higher successful treatment rates. Hence, the identification of treatment biomarkers may enhance translational and personalized medicine strategies, which in turn can shape the future for an improved quality of life of MDD patients.
Selected abbreviations and acronyms
| 2DE | = | two-dimensional gel electrophoresis |
| GC | = | gas chromatography |
| HPLC | = | high-performance liquid chromatography |
| iTRAQ | = | isobaric tags for relative and absolute quantitation |
| LC-MS | = | high-performance liquid chromatography-mass spectrometer |
| MDD | = | major depressive disorder |
| NMR | = | Nuclear magnetic resonance spectroscopy |
| PGDS | = | prostaglandin D2 synthase |
| PI3K-mTOR | = | phosphatidylincositol 3-kinase and the mammalian target of rapamycin |
| SDS | = | sodium dodecyl sulfate |
| SRM | = | selected reaction monitoring |
The author declares no conflict of interest and thanks Prof Chris Turck from the Max Planck Institute of Psychiatry, Prof Andrea Schmitt from the Department of Psychiatry and Psychotherapy of the LMU, and Prof Wagner Gattaz from the Institute of Psychiatry of the University of Sao Paulo. The author is funded by FAPESP (Sao Paulo Research Foundation, grant 2013/08711-3).
REFERENCES
- World Health Organization. Global burden of mental disorders and the need for a comprehensive, coordinated response from health and social sectors at the country level. Available at: http://apps.who.int/gb/ebwha/ pdf_files/EB130/B130_9-en.pdf. Updated December 1, 2011. Accessed January 7, 2014.
- MurroughJW.CharneyDS.Is there anything really novel on the antidepressant horizon?Curr Psychiatry Rep.20121464364922996298
- DiMatteoMR.LepperHS.CroghanTW.Depression is a risk factor for noncompliance with medical treatment: meta-analysis of the effects of anxiety and depression on patient adherence.Arch intern Med.20001602101210710904452
- Martins-de-SouzaD.Comprehending depression through proteomics.IntJ Neuropsychopharmacol.2012151373137422717368
- WilkinsMR.PasqualiC.AppelRD.et alFrom proteins to proteomes: large scale protein identification by two-dimensional electrophoresis and amino acid analysis.Biotechnology (N Y).19961461659636313
- O'FarrellPH.High resolution two-dimensional electrophoresis of proteins.J Biol Chern.197525040074021
- BjellqvistB.EkK.RighettiPG.et alIsoelectric focusing in immobilized pH gradients: principle, methodology and some applications.J Biochem Biophys Methods.198263173397142660
- GorgA.PostelW.GuntherS.et alApproach to stationary two-dimensional pattern: influence of focusing time and immobiline/carrier ampholytes concentrations.Electrophoresis.1988937462466646
- HubnerNC.RenS.MannM.Peptide separation with immobilized pi strips is an attractive alternative to in-gel protein digestion for proteome analysis.Proteomics.200884862487219003865
- WestermeierR.PostelW.WeserJ.GorgA.High-resolution two-dimensional electrophoresis with isoelectric focusing in immobilized pH gradients.J Biochem Biophys Methods.198383213306663005
- UnluM.MorganME.MindenJS.Difference gel electrophoresis: a single gel method for detecting changes in protein extracts.Electrophoresis.199718207120779420172
- LinkAJ.EngJ.SchieltzDM.et alDirect analysis of protein complexes using mass spectrometry.Nat Biotechnol.19991767668210404161
- FiliouMD.Martins-de-SouzaD.GuestPC.BahnS.TurckCW.To label or not to label: applications of quantitative proteomics in neuroscience research.Proteomics.20121273674722247077
- GygiSP.RistB.GerberSA.TurecekF.GelbMH.AebersoldR.Quantitative analysis of complex protein mixtures using isotope-coded affinity tags.Nat Biotechnol.19991799499910504701
- RossPL.HuangYN.MarcheseJN.et alMultiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents.Mol Cell Proteomics.200431154116915385600
- OngSE.BlagoevB.KratchmarovaI.et alStable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics.Mol Cell Proteomics.2002137638612118079
- Martins-de-SouzaD.GuestPC.Vanattou-SaifoudineN.HarrisLW.BahnS.Proteomic technologies for biomarker studies in psychiatry: advances and needs.Int Rev Neurobiol.2011101659422050849
- Martins-de-SouzaD.GuestPC.RahmouneH.BahnS.Proteomic approaches to unravel the complexity of schizophrenia.Exp Rev Proteomics.2012997108
- OliveiraBM.Martins-de-SouzaD.Large-scale analyses of schizophrenia proteome.Rev Psiq Clin.2013401619
- Martins-de-SouzaD.GuestPC.HarrisLW.et alIdentification of proteomic signatures associated with depression and psychotic depression in post-mortem brains from major depression patients.Transl Psychiatry.20122e8722832852
- Kaidanovich-BeilinO.ChaDS.MclntyreRS.Crosstalk between metabolic and neuropsychiatric disorders.F1000 Biol Rep.201241422802875
- RezinGT.AmboniG.ZugnoAl.QuevedoJ.StreckEL.Mitochondrial dysfunction and psychiatric disorders.Neurochem Res.2009341021102918979198
- StanleyJA.In vivo magnetic resonance spectroscopy and its application to neuropsychiatric disorders.Can J Psychiatry.20024731532612025430
- Martins-de-SouzaD.HarrisLW.GuestPC.BahnS.The role of energy metabolism dysfunction and oxidative stress in schizophrenia revealed by proteomics.Antioxid Redox Signal.2011152067207920673161
- FiliouMD.ZhangY.TeplytskaL.et alProteomics and metabolomics analysis of a trait anxiety mouse model reveals divergent mitochondrial pathways.Biol Psychiatry.2011701074108221791337
- VaradarajuluJ.SchmittA.FalkaiP.AlsaifM.TurckCW.Martins-deSouzaD.Differential expression of HINT1 in schizophrenia brain tissue.Eur Arch Psychiatry Clin Neurosci.201226216717221553311
- BarbierE.WangJB.Anti-depressant and anxiolytic like behaviors in PKCI/HINT1 knockout mice associated with elevated plasma corticosterone level.BMC Neurosci.20091013219912621
- VaradarajuluJ.LebarM.KrishnamoorthyG.et alIncreased anxietyrelated behaviour in HINT1 knockout mice.Behav Brain Res.201122030531121316396
- Martins-de-SouzaD.GuestPC.Vanattou-SaifoudineN.RahmouneH.BahnS.Phosphoproteomic differences in major depressive disorder postmortem brains indicate effects on synaptic function.Eur Arch Psychiatry Clin Neurosci.201226265766622350622
- Johnston-WilsonNL.SimsCD.HofmannJP.et alDisease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. The Stanley Neuropathology Consortium.Mol Psychiatry.2000514214910822341
- BeasleyCL.PenningtonK.BehanA.WaitR.DunnMJ.CotterD.Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: evidence for disease-associated changes.Proteomics.200663414342516637010
- HensleyK.VenkovaK.ChristovA.GunningW.ParkJ.Collapsin response mediator protein-2: an emerging pathologic feature and therapeutic target for neurodisease indications.Mol Neurobiol.20114318019121271304
- HuangJT.LewekeFM.OxleyD.et alDisease biomarkers in cerebrospinal fluid of patients with first-onset psychosis.PLoS Med.20063e42817090210
- DitzenC.TangN.JastorffAM.et alCerebrospinal fluid biomarkers for major depression confirm relevance of associated pathophysiology.Neuropsychopharmacology.2012371013102522169944
- Martins-De-SouzaD.WobrockT.ZerrI.et alDifferent apolipoprotein E, apolipoprotein A1 and prostaglandin-H2 D-isomerase levels in cerebrospinal fluid of schizophrenia patients and healthy controls.World J Biol Psychiatry.20101171972820446881
- CarboniL.BecchiS.PiubelliC.et alEarly-life stress and antidepressants modulate peripheral biomarkers in a gene-environment rat model of depression.Prog Neuropsychopharmacoi Biol Psychiatry.20103410371048
- ZhangY.FiliouMD.ReckowS.et alProteomic and metabolomic profiling of a trait anxiety mouse model implicate affected pathways.Mol Cell Proteomics.201110M111.008110
- XuHB.ZhangRF.LuoD.et alComparative proteomic analysis of plasma from major depressive patients: identification of proteins associated with lipid metabolism and immunoregulation.Int J Neuropsychopharmacol.2012151413142522717272
- Martins-de-SouzaD.Is the word' biomarker' being properly used by proteomics research in neuroscience?Eur Arch Psychiatry Clin Neurosci.201026056156220155362
- Martins-de-SouzaD.MaccarroneG.IsingM.et alBlood mononuclear cell proteome suggests Integrin and Ras signaling as critical pathways for antidepressant treatment response.Biol Psychiatry. 2014. In press.
- RaamsdonkLM.TeusinkB.BroadhurstD.et alA functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations.Nat Biotechnol.200119455011135551
- NicholsonJK.LindonJC.HolmesE.'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data.Xenobiotica.1999291181118910598751
- Kaddurah-DaoukR.KrishnanKR.Metabolomics: a global biochemical approach to the study of central nervous system diseases.Neuropsychopharmacology.20093417318618843269
- ShinMH.Lee doY.LiuKH.FiehnO.KimKH.Evaluation of sampling and extraction methodologies for the global metabolic profiling of Saccharophagus degradans.Anal Chem.2010826660666620669998
- CubbonS.AntonioC.WilsonJ.Thomas-OatesJ.Metabolomic applications of HILIC-LC-MS.Mass Spectrom Rev.20102967168419557839
- RojoD.BarbasC.RuperezFJ.LC-MS metabolomics of polar compounds.Bioanalysis.201241235124322651567
- NemutluE.ZhangS.JuranicNO.TerzicA.MacuraS.DzejaP.180-assisted dynamic metabolomics for individualized diagnostics and treatment of human diseases.Croat Med J.20125352953423275318
- SmolinskaA.BlanchetL.BuydensLM.WijmengaSS.NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review.Anal Chim Acta.2012750829723062430
- ReoNV.NMR-based metabolomics.Drug Chem Toxicol.20022537538212378948
- HaradaK.FukusakiE.BambaT.SatoF.KobayashiA.In vivo 15N-enrichment of metabolites in suspension cultured cells and its application to metabolomics.Biotechnol Prog.2006221003101116889377
- ZamboniN.SauerU.Novel biological insights through metabolomics and 13C-flux analysis.Curr Opin Microbiol.20091255355819744879
- BeckerS.KortzL.HelmschrodtC.ThieryJ.CeglarekU.LC-MS-based metabolom ics in the clinical laboratory.J Chromatogr B Analyt Technol Biomed LifeSci.2012883-8846875
- DettmerK.AronovPA.HammockBD.Mass spectrometry-based metabolomics.Mass Spectrom Rev.200726517816921475
- MilneSB.MathewsTP.MyersDS.IvanovaPT.BrownHA.Sum of the parts: mass spectrometry-based metabolomics.Biochemistry.2013523829384023442130
- GarciaA.BarbasC.Gas chromatography-mass spectrometry (GC-MS)based metabolomics.Methods Mol Biol.201170819120421207291
- ZhouB.XiaoJF.TuliL.RessomHW.LC-MS-based metabolomics.Mol Biosyst.2012847048122041788
- RamautarR.SomsenGW.de JongGJ.CE-MS for metabolomics: developments and applications in the period 2010-2012.Electrophoresis.201334869823161106
- LeiZ.HuhmanDV.SumnerLW.Mass spectrometry strategies in metabolomics.J Biol Chem.2011286254352544221632543
- DaiY.LiZ.XueL.et alMetabolomics study on the anti-depression effect of xiaoyaosan on rat model of chronic unpredictable mild stress.J Ethnopharmacol.201012848248920079416
- ZhengS.YuM.LuX.et alUrinary metabonomic study on biochemical changes in chronic unpredictable mild stress model of depression.Clin Chim Acta.201041120420919913000
- ZhangF.JiaZ.GaoP.et alMetabonomics study of urine and plasma in depression and excess fatigue rats by ultra fast liquid chromatography coupled with ion trap-time of flight mass spectrometry.Mol Biosyst.2010685286120567771
- WangX.ZengC.LinJ.et alMetabonomics approach to assessing the modulatory effects of St John's wort, ginsenosides, and clomipramine in experimental depression.J Proteome Res.2012116223623023110693
- Kaddurah-DaoukR.YuanP.BoyleSH.et alCerebrospinal fluid metabolome in mood disorders-remission state has a unique metabolic profile.Sci Rep.2012266722993692
- PaigeLA.MitchellMW.KrishnanKR.Kaddurah-DaoukR.SteffensDC.A preliminary metabolomic analysis of older adults with and without depression.Int J Geriatr Psychiatry.20072241842317048218
- SteffensDC.WeiJ.KrishnanKR.et alMetabolomic differences in heart failure patients with and without major depression.J Geriatr Psychiatry Neurol.20102313814620101071
- ZhengP.GaoHC.LiQ.et alPlasma metabonomics as a novel diagnostic approach for major depressive disorder.J Proteome Res.2012111741174822239730
- ZhengP.WangY.ChenL.et alIdentification and validation of urinary metabolite biomarkers for major depressive disorder.Mol Cell Proteomics.20131220721423111923
- Kaddurah-DaoukR.BoyleSH.MatsonW.et alPretreatment metabotype as a predictor of response to sertraline or placebo in depressed outpatients: a proof of concept.Transl Psychiatry.20111.piie26
- Kaddurah-DaoukR.BogdanovMB.WikoffWR.et alPharmacometabolomic mapping of early biochemical changes induced by sertraline and placebo.Transl Psychiatry.20133e22323340506
- Martins-de-SouzaD.Translational strategies to schizophrenia from a proteomic perspective.Transl Neurosci.20123300302
- CoulombeB.Mapping the disease protein interactome: toward a molecular medicine GPS to accelerate drug and biomarker discovery.J Proteome Res.20111012012520939596
- WilliamsonMP.SutcllffeMJ.Protein-protein interactions.Biochem Soc Trans.20103887587820658969
- ChenJ.ZhouJ.SandersCK.NolanJP.CaiH.A surface display yeast twohybrid screening system for high-throughput protein interactome mapping.Anal Biochem.2009390293719298787
- ScifoE.SzwajdaA.DebskiJ.et alDrafting the CLN3 protein interactome in SH-SY5Y human neuroblastoma cells: a label-free quantitative proteomics approach.J Proteome Res.2013122101211523464991
- Van LeeneJ.StalsH.EeckhoutD.et alA tandem affinity purificationbased technology platform to study the cell cycle interactome in Arabidopsis thaliana.Mol Cell Proteomics.200761226123817426018
- MarkhamK.BaiY.Schmitt-UlmsG.Co-immunoprecipitations revisited: an update on experimental concepts and their implementation for sensitive interactome investigations of endogenous proteins.Anal Bioanal Chem.200738946147317583802
- BonfiglioJJ.MaccarroneG.RewertsC.et alCharacterization of the B-Raf interactome in mouse hippocampal neuronal cells.J Proteomics.20117418619821055488
- NikolskyY.NikolskayaT.BugrimA.Biological networks and analysis of experimental data in drug discovery.Drug Discov Today.20051065366215894230
- SteinA.MoscaR.AloyP.Three-dimensional modeling of protein interactions and complexes is going'omics.Curr Opin Struct Biol.20112120020821320770
- ValenciaA.PazosF.Computational methods for the prediction of protein interactions.Curr Opin Struct Biol.20021236837312127457
- WeichhartT.SaemannMD.The P13K/Akt/mTOR pathway in innate immune cells: emerging therapeutic applications.Ann Rheum Dis.200867(suppl 3)iii707419022819
- Pilot-StorckF.ChopinE.RualJF.et alInteractome mapping of the phosphatidylinositol 3-kinase-mammalian target of rapamycin pathway identifies deformed epidermal autoregulatory factor-1 as a new glycogen synthase kinase-3 interactor.Mol Cell Proteomics.201091578159320368287
- JiaP.KaoCF.KuoPH.ZhaoZ.A comprehensive network and pathway analysis of candidate genes in major depressive disorder.BMC Syst Biol.20115 Suppl 3S1222784618
- Sperner-UnterwegerB.KohlC.FuchsD.Immune changes and neurotransmitters: Possible interactions in depression?Prog Neuropsychopharmacol Biol Psychiatry.20144826827623085509
- Martins-de-SouzaD.Proteomics is not only a biomarker discovery tool.Proteomics Clin Appl.2009311361139
- FranceschiniA.SzklarczykD.FrankildS.et alSTRING v9.1: protein-protein interaction networks, with increased coverage and integration.Nucleic Acids Res.201341 (Database issue)D80881523203871
- SmootME.OnoK.RuscheinskiJ.WangPL.IdekerT.Cytoscape 2.8: new features for data integration and network visualization.Bioinformatics.20112743143221149340
- SchrattenholzA.GroebeK.SoskicV.Systems biology approaches and tools for analysis of interactomes and multi-target drugs.Methods Mol Biol.2010662295820824465
- TurckCW.IrisF.Proteome-based pathway modelling of psychiatric disorders.Pharmacopsychiatry.201144 (suppl 1)55461
- FiliouMD.TurckCW.Martins-de-SouzaD.Quantitative proteomics for investigating psychiatric disorders.Proteomics Clin Appl.20115384921280236
- GormannsP.MuellerNS.DitzenC.WolfS.HolsboerF.TurckCW.Phenome-transcriptome correlation unravels anxiety and depression related pathways.J Psychiatr Res.20114597397921255794
- YangY.YangD.TangG.et alProteomics reveals energy and glutathione metabolic dysregulation in the prefrontal cortex of a rat model of depression.Neuroscience.201324719120023727007
- BisgaardCF.BakS.ChristensenT.JensenON.EnghildJJ.WiborgO.Vesicular signalling and immune modulation as hedonic fingerprints: proteomic profiling in the chronic mild stress depression model.J Psychopharmacol.2012261569158323139383
- XuZ.HouB.ZhangY.et alAntidepressive behaviors induced byenriched environment might be modulated by glucocorticoid levels.Eur Neuropsychopharmacoi.200919868875
- CarboniL.Peripheral biomarkers in animal models of major depressive disorder.Dis Markers.2013358
- ConnollyKR.ThaseME.Emerging drugs for major depressive disorder.Expert Opin Emerg Drugs.20121710512622339643
- Martins-de-SouzaD.Biomarkers for psychiatric disorders: where are we standing?Dis Markers.2013351224167343
- DomeniciE.WilleDR.TozziF.et alPlasma protein biomarkers for depression and schizophrenia by multi analyte profiling of case-control collections.PLoS One.20105e916620161799
- RaedlerTJ.WiedemannK.CSF-studies in neuropsychiatric disorders.Neuro Endocrinol Lett.20062729730516807525
- BahnS.SchwarzE.HarrisLW.Martins-de-SouzaD.RahmouneH.GuestPC.Biomarker blood tests for diagnosis and management of mental disorders: focus on schizophrenia.Rev Psiq Clin.2013407

