Abstract
The purine degradation pathway in humans ends with uric acid, which has low water solubility. When the production of uric acid is increased either by elevated purine intake or by impaired kidney function, uric acid will accumulate in the blood (hyperuricemia). This increases the risk of gout, a disease described in humans for at least 1000 years. Many lower organisms, such as the yeast Arxula adeninivorans, possess the enzyme, urate oxidase that converts uric acid to 5-hydroxyisourate, thus preventing uric acid accumulation. We have examined the complete purine degradation pathway in A. adeninivorans and analyzed enzymes involved. Recombinant adenine deaminase, guanine deaminase, urate oxidase and endogenous xanthine oxidoreductase have been investigated as potential additives to degrade purines in the food. Here, we review the current model of the purine degradation pathway of A. adeninivorans and present an overview of proposed enzyme system with perspectives for its further development.
Abbreviations
| HIU | = | 5-hydroxyisourate |
| OHCU | = | 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline |
| dcw | = | dry cell weight |
Introduction
Foods of animal and vegetable origin contain different amounts of RNA, DNA, nucleotides, nucleosides and free purines. These complex compounds from exogenous sources as well as from cell turnover are broken down via free purine bases to uric acid, the end product of the human purine degradation pathway.Citation1 Approximately 80% of uric acid excretion is through the kidneys and impairment of their function and/or elevation of uric acid production leads to hyperuricemia – a condition characterized by a concentration serum urate above its saturation level.Citation2,3 Hyperuricemia is a risk factor for gout,Citation4 an inflammatory arthritis caused by monosodium urate crystals forming in joints.Citation5 The disease is characterized by excruciating pain, swelling and warmth of the affected joints called a gout attack.Citation6 Prevalence of gout is increasing in the population worldwide and it is believed to be caused by rising obesity, diet rich in purines and alcohol consumption.Citation4
Treatment of gout depends on the clinical phase of the disease and the patient's specific risk factors (high serum urate, body weight, alcohol consumption).Citation7 Acute gout attacks are treated with non-steroidal anti-inflammatory drugs (NSAIDs), colchicine or in rare cases with corticosteroids and interleukin-1β inhibitors. The goal of the treatment at this stage is pain relief and elimination of inflammation in the joints. During the second stage (between the attacks), drugs lowering urate concentration are given to the patient (uricostatics, uricosurics or uricolytics), which help to prevent future attacks.Citation7-9
Despite various pharmaceutical options, a reduction in intake of purine-rich foods is recommended as a basis of gout therapy.Citation3 Patients should avoid eating meat and organ meats, seafood, legumes and other high-purine-content food. The intake of alcohol should also be restricted. These dietary changes are essential and enhance the effectiveness of every pharmaceutical treatment.Citation3,4
To provide patients with better control of the disease with reduced symptoms and frequency of gout attacks, a novel enzyme system decreasing the purine concentration of food products has been developed.Citation10-13 This system is an enzyme mixture that, at present, consists of 4 purine-degrading enzymes: adenine deaminase, guanine deaminase, xanthine oxidoreductase and urate oxidase, which simultaneously break down purines to a water-soluble 5-hydroxyisourate. The goal of this article is to give an overview of the composition and application of this enzyme system.
Enzymes and Their Role in the Purine Catabolism in A. adeninivorans
The studies of purine degradation pathway in various organisms have shown that this process can end at different stages in the enzymatic degradation pathway, and thus leads to an excretion of various end products.Citation14,15 The analysis of A. adeninivorans genome revealed a presence of almost all necessary genes essential for purine degradation to CO2 and NH3 ().Citation16
Figure 1. Schema of proposed purine degradation pathway in A. adeninivorans. The presence of OHCU decarboxylase has not been confirmed yet.
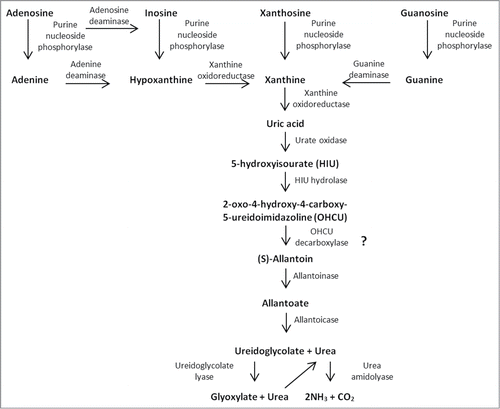
The degradation starts with the 5’-nucleotidase that hydrolyses purine nucleotides (AMP, GMP, IMP and XMP) to nucleosides and phosphate. Adenosine, inosine, xanthosine and guanosine are then cleaved with purine nucleoside phosphorylase to free purine bases (adenine, hypoxanthine, xanthine and guanine) and ribose-1-phosphate. Adenine and guanine are deaminated to hypoxanthine and xanthine, respectively, by the enzymes, adenine deaminase and guanine deaminase. Hypoxanthine and xanthine are both substrates for xanthine oxidoreductase, which oxidises them to uric acid, the last common product of purine degradation in all organisms and the final product in humans. Further degradation of uric acid varies from species to species.Citation15
In most organisms, except for primates, reptiles, birds and terrestrial insects, uric acid undergoes degradation to allantoin. Kahn and TiptonCitation17 suggested that allantoin is not the true product of the urate oxidase oxidation in soybean and further investigation led to the identification of 2 new enzymes that participate in formation of allantoin. The complete pathway of uric acid degradation involves urate oxidase, which produces 5-hydroxyisourate (HIU); 5-hydroxyisourate hydrolase (HIU hydrolase), which yields 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU); and OHCU decarboxylase that leads to formation of (S)-allantoin and CO2.Citation18
The sequence analysis of A. adeninivorans genome did reveal the presence of HIU hydrolase but it is not known whether the OHCU produced by this enzyme is degraded enzymatically or degrades spontaneously, because the appropriate gene has not be found to date.
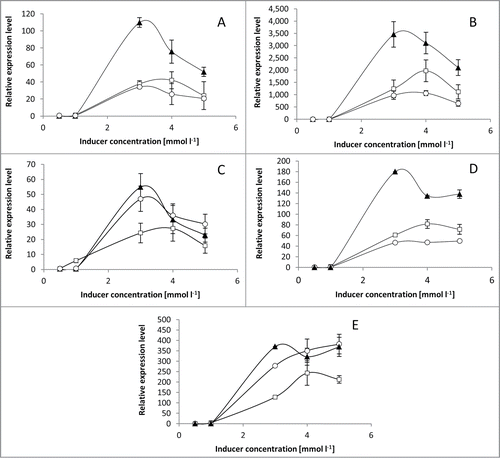
All of the characterized enzymes, adenine and guanine deaminases, xanthine oxidoreductase and urate oxidase have been shown by qPCR and enzyme analysis to be inducible.Citation10-13 The expression of all genes was monitored over time with different concentration of inducers using SYBR Green fluorescent dye for double-stranded DNA quantification. The mRNA accumulation was strongly enhanced by the presence of hypoxanthine and adenine in cultivation medium (2 times higher mRNA level for hypoxanthine than adenine in case of adenine deaminase, guanine deaminase and xanthine oxidoreductase); uric acid also strongly induced the urate oxidase gene (). The results are in agreement with measured enzyme activities, which were highest when the hypoxanthine was present in the cultivation medium (data not shown). Only in case of urate oxidase was the induction of its gene equally good with adenine as it was with hypoxanthine.Citation10-13 Clearly, the mRNA relation between every inducer is reflected here in the enzyme activity, however, the amount of accumulated transcript does not translate into the amount protein synthesized. The very high (up to 3,500, ) relative expression level of xanthine oxidoreductase gene was not related with the actual protein accumulation. However, as shown by Vogel and MarcotteCitation19 the mRNA level corresponds to the level of protein in only around 40%, the remaining 60% being dependent on post-transcriptional, translational and degradation regulation.
Figure 2. The influence of nitrogen source on transcript accumulation of (A) AADA, (B) AXOR, (C) AUAY (D) AGDA, (E) AUOR. The A. adeninivorans cells were cultivated in yeast minimal medium with glucose (1%) as carbon source and various purines as nitrogen sources at different concentrations for 24 h. The transcript was analyzed by qPCR. Used markers: (□) adenine, (▴) hypoxanthine, (○) uric acid.
These and other genes coding for remaining enzymes from purine degradation pathway were recently analyzed for adenine inducibility using microarray technology.Citation16 The results demonstrated that adenine positively influences expression of all the genes in the pathway except the urea amidolyase gene. Adenine however, does not directly induce the genes. It has been shown using a strain lacking xanthine oxidoreductase activity that the true inducer of urate oxidase gene is in fact uric acid. Thus adenine, as well as other purines, must be first converted to uric acid before they can act as an inducer.Citation11 An analogous situation may apply to other enzymes of the pathway (allantoinase, allantoicase, ureidoglycolate hydrolase), as is the in case of Aspergillus nidulans and Neurospora crassaCitation20, 21 where all genes are activated with uric acid. The true inducers of A. adeninivorans enzymes of the pathway will be tested once appropriate knockout mutants are available.
It has been shown in A. nidulans, that the purine degradation pathway is regulated by a UaY transcription factor, which mediates uric acid induction. A blast search of A. adeninivorans genome shows that the AUAY gene has been identified which has similarity to the positive regulators of purine utilization. Like the positive regulators it belongs to the zinc finger transcription factors, which have a ZnII2Cys6 DNA binding motif. This class of binuclear zinc proteins is found only in fungal kingdom and is involved in the regulation of many pathways.Citation22 The best-known and first-described protein of this class is Gal4p.Citation23 The inducibility of A. adeninivorans AUAY gene was analyzed by qPCR in yeast samples grown on adenine, hypoxanthine and uric acid as nitrogen sources. It was shown that uric acid functions as an inducer almost as well as hypoxanthine, whereas the adenine induction effect is about half that of hypoxanthine (at 3 mM). Again, it is possible that the only “true” inducer of this transcription factor is uric acid as in case of urate oxidase (AUOR) gene. The inducibility of all the characterized enzymes of purine degradation pathway in A. adeninivorans is presented in . The same induction pattern can be observed for all genes suggesting that it can be modulated by the same transcription factor, as shown for A. nidulans.Citation22
Production of Low-Purine Content Food with use of A. adeninivorans
A. adeninivorans belongs to the phylum Ascomycetes and exhibits some unusual properties. The yeast is tolerant to high salt concentration (up to 20% NaCl) and to high temperatures (grows up to 48°C). As a dimorphic organism, it undergoes a morphological change from budding cells to a mycelial form when cultivated above 42°C.Citation24 Moreover, A. adeninivorans can use an extraordinarily large number of C and N sources as n-alkanes, starch, polyalcohols and organic acids, tannins and tannic acid as a carbon source and nitrate as nitrogen source.Citation25-27 In addition, this yeast can grow on purines, such as uric acid, adenine and hypoxanthine as a carbon and/or nitrogen source.Citation28 Due to its robustness, outstanding growth parameters and broad C-and N-source spectrum, A. adeninivorans is an ideal organism for various biotechnological applications.
The aim of this project was to develop a method to degrade purines in food products in order to prevent hyperuricemia. From preliminary investigations it was known that A. adeninivorans LS3 grows on purines as the sole N-source and the genome sequence analysis revealed presence of all genes from purine degradation pathway. The products of those genes, adenine deaminase (Aadap), guanine deaminase (Agdap) and urate oxidase (Auoxp), were successfully synthesized in transgenic A. adeninivorans cells using the Xplor®2 transformation/expression platform as described by Jankowska et al.Citation10 and Trautwein-Schult et al.Citation12,13 The productivity of each transformant strain was optimized by modification of the medium, temperature and carbon source concentration. The highest productivity for guanine deaminase and urate oxidase was obtained in yeast minimal medium supplemented with 1% glucose after 24 h of cultivation at 30°C whereas for adenine deaminase the conditions were yeast minimal medium with 2% glucose for 5 h at 37°C. Eventually, the activity attained was up to 15,000 U g‑1 dcw (dry cell weight) for adenine deaminase, up to 73 U g‑1 dcw for guanine deaminase and up to 25 U g‑1 dcw for urate oxidase. However transformants exhibiting recombinant xanthine oxidoreductase (Axorp) activity have not yet been obtained. The cultivation procedure for this enzyme in the wild-type strain was also optimized; yeast minimal medium with 1% glucose and 2.5 mM hypoxanthine, 5 h of shaking at 30°C. The yield attained was 22 U gCitation‑1 dcw, which is similar to the yield for urate oxidase.Citation11
All four enzymes have been purified and biochemically characterized. The cells were first mechanically disrupted to release intracellular proteins. Recombinant adenine deaminase, guanine deaminase and urate oxidase all had a sequence coding for His-tag at the C-terminus of the gene which enabled purification using Ni2+-charged His-bind resin. The wild-type xanthine oxidoreductase was partially purified on an ion exchange column followed by the size exclusion chromatography. The mass analysis of the purified enzymes in the native and denaturing conditions confirmed the monomeric structure of both deaminases and dimeric structure of xanthine oxidoreductase and urate oxidase, that were assumed based on the gene sequence.
The optimal pH was determined by measuring enzyme activity in different buffers in the pH range of 2–12. For determining the optimal temperature, the enzymes and other assay solution were equilibrated in chosen temperatures for 5 min before start of the reaction. The pH and thermal stabilities were assessed after incubation of enzymes under given conditions for set times (described by Jankowska et al.,Citation10,11 Trautwein-Schult et al.Citation12,13). The optimal assay conditions and pH and temperature stabilities were similar enough to enable the enzymes to work well when mixed in a single solvent of fixed pH and temperature (pH 8.0 and 40°C – ).
Table 1. Comparison of the properties of purified recombinant adenine deaminase (Aada6hp), guanine deaminase (Agda6hp), urate oxidase (Auor6hp) and wild-type xanthine oxidoreductase (Axorp)
Three of 4 enzymes are highly specific for their substrates, whereas xanthine oxidoreductase is more versatile and can oxidase other purines and purine analogs using NAD+ as an electron acceptor. All KM values for the natural substrates are in the micromolar range starting with 26 μM for guanine (with guanine deaminase) to 660 μM for adenine (with adenine deaminase). The turnover number (kcat) is the lowest for recombinant guanine deaminase, 1.5 s−1, and increases in adenine deaminase - 20.8 s−1, and urate oxidase - 121 s−1. This constant could not be determined for xanthine oxidoreductase because of low purity of the wild-type enzyme.
Application of Enzymes for the Production of Low-Purine Content Food
Initial experiments to test the functionality of an enzyme mixture (adenine deaminase, guanine deaminase, xanthine oxidoreductase and urate oxidase) were carried out on a solution containing equal amounts of xanthine, hypoxanthine, adenine, guanine and uric acid. For all purines a significant reduction in concentration was detected.
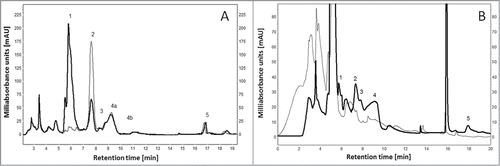
For tests with beef broth and yeast extract, both constituents of a variety of processed foods, single enzymes or enzyme mixes were applied. Adenine deaminase and urate oxidase were able to completely remove adenine and uric acid from beef broth when added as sole enzyme.Citation10,12 Guanine deaminase and xanthine oxidoreductase were tested in a mixture with the remaining 2 enzymes on both beef broth and yeast extract leading to a substantial decrease of purine concentration in those products ().Citation11,13
Figure 3. Production of food with low purine content using beef broth as an example. Presented is the profile of the beef broth before (thick line) and after (thin line) enzyme treatment with (A) 4.2 U adenine deaminase (Aadap) for 20 min and (B) an enzyme mixture containing 1 U of xanthine dehydrogenase (Axorp) and 0.125 U of adenine deaminase (Aadap), guanine deaminase (Agdap) and urate oxidase (Auorp) respectively plus 3 mg ml−1 NAD+ for 240 min. Peaks: 1 – adenine, 2 – hypoxanthine, 3 – xanthine, 4a,b – guanine, 5 – uric acid.
The results of analysis of rolled fillet of ham are shown in . The untreated food contained 21.1 mg/l guanine and smaller amounts of hypoxanthine (9.4 mg/l), adenine (1.4 mg/l), xanthine (1.1 mg/l). Uric acid was not detected. The incubation of dissolved rolled fillet of ham over 240 min with the enzyme mixture (0.2 U/ml adenine deaminase (Aadap) 0.2 U/ml guanine deaminase (Agdap), 0.2 U/ml urate oxidase (Auorp) and 0.2 U/ml xanthine oxidoreductase (Axorp)) caused a reduction in each of the purines.
Table 2. Production of rolled fillet of ham with low purine content. A rolled fillet of macerated ham was incubated with the enzyme mixture consisting of adenine deaminase (Aadap), guanine deaminase (Agdap), xanthine dehydrogenase (Axorp) and urate oxidase (Auorp). The purine contents were analyzed by HPLC
Conclusions and Further Perspectives
The present approach for reduction of purine content in food consists of 4 enzymes, of which 3 were recombinant. The experiments with those enzymes performed on food constituents provided evidence of the potential of the approach and support the value of further work on the system.
The next step already undertaken to expand the range of the system to nucleosides and products of urate oxidase action is production of 2 additional enzymes. Purine nucleoside phosphorylase will hydrolyse adenosine, inosine, xanthosine and guanosine and provide substrates for already existing enzymes, and 5-hydroxyisourate hydrolase will process 5-hydroxyisourate, which is synthesized from uric acid.
Once all enzymes are available, their yield will have to be increased. All cell cultivations in the described experiments were carried out in shake flasks as a batch culture, however, it has been shown for tannase produced in A. adeninivoransCitation29 that continuous cultivation in a fermenter results in a higher cell density and thus a more efficient protein production. The over-expression and/or modification of the transcription factors (e.g., UaY) can also provide a higher yield, resistance to stress and promote the cell growth without production of new, unwanted compounds.
For a wide application of the enzymes in the food industry, it is necessary to develop a method by which the foods are treated prior to consumption. The food could be depleted of purines during the production process in the factory; however a more convenient way would be an addition of enzymes to spice mixtures which are applied to food during preparation. Another possibility is to produce encapsulated enzymes to be consumed before a meal which then act on the food during digestion in the intestines. Two similar projects aiming in lowering purine content in the food have been reported in the literature to date. In 1978, Sakai and JunCitation30 proposed to use Pseudomonas synxantha as an enzyme producer for food industry. However the progress of this approach is not known. In 2013 Cheng et al.Citation31 presented an alternative approach using a common inhabitant of human intestinal tract, Lactobacillus bulgaria. The urate oxidase gene from Candida utilis was cloned into the L. bulgaria with the intention to colonize human gut with this bacterium in order to degrade uric acid before it reaches the blood stream. However further work on this approach has not been published.
Depending on the chosen mode of action, every application requires the enzymes to be in an appropriate form that preserve their activity and allow for storage for long periods. Thus, before the system to hydrolyse purines in food products using A. adeninivorans will be commercially available, further research must be done to develop a commercially relevant system. The most challenging problem will be to produce the enzymes in a form that preserves their activity in a way that is compatible with food and allows for storage for long periods.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to Dr. M Giersberg for helpful discussions and for critical reading of the manuscript.
Funding
The research work was supported by grant from the BMWi (Project No. KF2131601MD8).
References
- Burkard M, Huth K. Hyperurikämie und Gicht. Ernährung und Fasten als Therapie. Berlin: Springer, 2010. 255-270. ISBN-13 978-3-540-88809-3
- Masseoud D, Rott K, Liu-Bryan R, Agudelo C. Overview of hyperuricaemia and gout. Curr Pharm Des 2005; 11:4117-24; PMID:16375732; http://dx.doi.org/10.2174/138161205774913318
- Schlesinger N. Dietary factors and hyperuricaemia. Curr Pharm Des 2005; 11:4133-8; PMID:16375734; http://dx.doi.org/10.2174/138161205774913273
- Saag KG, Choi H. Epidemiology, risk factors, and lifestyle modifications for gout. Arthritis Res Therapy 2006; 8 Suppl 1:S2; PMID:16820041; http://dx.doi.org/10.1186/ar1907
- Terkeltaub R. Update on gout: new therapeutic strategies and options. Nat Rev Rheumatol 2010; 6:30-8; PMID:20046204; http://dx.doi.org/10.1038/nrrheum.2009.236
- Mandell BF. Clinical manifestations of hyperuricemia and gout. Cleveland Clinic J Med 2008; 75 Suppl 5:S5-8; http://dx.doi.org/10.3949/ccjm.75.Suppl_5.S5
- Eggebeen AT. Gout: an update. Am Fam Physician 2007; 76:801-8; PMID:17910294
- Burns CM, Wortmann RL. Gout therapeutics: new drugs for an old disease. Lancet 2011; 377:165-77; PMID:20719377; http://dx.doi.org/10.1016/S0140-6736(10)60665-4
- Schlesinger N, Dalbeth N, Perez-Ruiz F. Gout–what are the treatment options? Expert Opin Pharmacother 2009; 10:1319-28; PMID:19463070; http://dx.doi.org/10.1517/14656560902950742
- Jankowska DA, Faulwasser K, Trautwein-Schult A, Cordes A, Hoferichter P, Klein C, Bode R, Baronian K, Kunze G. Arxula adeninivorans recombinant adenine deaminase and its application in the production of food with low purine content. J Appl Microbiol 2013; 115:1134-46; PMID:23902582; http://dx.doi.org/10.1111/jam.12317
- Jankowska DA, Trautwein-Schult A, Cordes A, Hoferichter P, Klein C, Bode R, Baronian K, Kunze G. Arxula adeninivorans xanthine oxidoreductase and its application in the production of food with low purine content. J Appl Microbiol 2013; 115:796-807; PMID:23773263; http://dx.doi.org/10.1111/jam.12284
- Trautwein-Schult A, Jankowska D, Cordes A, Hoferichter P, Klein C, Matros A, Mock HP, Baronian K, Bode R, Kunze G. Arxula adeninivorans recombinant urate oxidase and its application in the production of food with low uric acid content. J Mol Microbiol Biotechnol 2013; 23:418-30; PMID:24022585; http://dx.doi.org/10.1159/000353847
- Trautwein-Schult A, Jankowska D, Cordes A, Hoferichter P, Klein C, Matros A, Mock HP, Baronian K, Bode R, Kunze G. Arxula adeninivorans recombinant guanine deaminase and its application in the production of food with low purine content. J Mol Microbiol Biotechnol 2014; 24:67-81; PMID:24481069; http://dx.doi.org/10.1159/000357674
- Scazzocchio C. The purine degradation pathway, genetics, biochemistry and regulation. Prog Ind Microbiol 1994; 29:221-57; PMID:7765126
- Vogels GD, Van der Drift C. Degradation of purines and pyrimidines by microorganisms. Bacteriol Rev 1976; 40:403-68; PMID:786256
- Kunze G, Gaillardin C, Czernicka M, Durrens P, Martin T, Böer E, Gabaldón T, Cruz J, Talla E, Marck C, et al. The complete genome of Blastobotrys (Arxula) adeninivorans LS3 – a yeast of biotechnological interest. Nat Biotechnol 2014; 7:66; PMID:24834124; http://dx.doi.org/10.1186/1754-6834-7-66
- Kahn K, Tipton PA. Kinetic mechanism and cofactor content of soybean root nodule urate oxidase. Biochemistry 1997; 36:4731-8; PMID:9125493; http://dx.doi.org/10.1021/bi963184w
- Cendron L, Berni R, Folli C, Ramazzina I, Percudani R, Zanotti G. The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation. J Biol Chem 2007; 282:18182-9; PMID:17428786; http://dx.doi.org/10.1074/jbc.M701297200
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012; 13:227-32; PMID:22411467
- Scazzocchio C, Darlington AJ. The induction and repression of the enzymes of purine breakdown in Aspergillus nidulans. Biochim Biophys Acta 1968; 166:557-68; PMID:5680610; http://dx.doi.org/10.1016/0005-2787(68)90243-8
- Griffith AB, Garrett RH. Xanthine dehydrogenase expression in Neurospora crassa does not require a functional nit-2 regulatory gene. Biochem Genet 1988; 26:37-52; PMID:2967694; http://dx.doi.org/10.1007/BF00555487
- Cecchetto G, Richero M, Oestreicher N, Muro-Pastor MI, Pantano S, Scazzocchio C. Mutations in the basic loop of the Zn binuclear cluster of the UaY transcriptional activator suppress mutations in the dimerisation domain. Fungal Genet Biol 2012; 49:731-43; PMID:22760060; http://dx.doi.org/10.1016/j.fgb.2012.06.009
- MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 2006; 70:583-604; PMID:16959962; http://dx.doi.org/10.1128/MMBR.00015-06
- Wartmann T, Kruger A, Adler K, Duc BM, Kunze I, Kunze G. Temperature-dependent dimorphism of the yeast Arxula adeninivorans LS3. Antonie Van Leeuwenhoek 1995; 68:215-23; PMID:8572679; http://dx.doi.org/10.1007/BF00871818
- Böer E, Bode R, Mock HP, Piontek M, Kunze G. Atan1p-an extracellular tannase from the dimorphic yeast Arxula adeninivorans: molecular cloning of the ATAN1 gene and characterization of the recombinant enzyme. Yeast 2009; 26:323-37; PMID:19387973; http://dx.doi.org/10.1002/yea.1669
- Middelhoven WJ, De Kievit H, Biesbroek AL. Yeast species utilizing uric acid, adenine, n-alkylamines or diamines as sole source of carbon and energy. Antonie Van Leeuwenhoek 1985; 51:289-301; PMID:4091535; http://dx.doi.org/10.1007/BF02439938
- Middelhoven WJ, de Jong IM, de Winter M. Arxula adeninivorans, a yeast assimilating many nitrogenous and aromatic compounds. Antonie Van Leeuwenhoek 1991; 59:129-37; PMID:1854187; http://dx.doi.org/10.1007/BF00445657
- Middelhoven WJ, Hoogkamer-Te Niet MC, Kreger-Van Rij NJ. Trichosporon adeninovorans sp. nov., a yeast species utilizing adenine, xanthine, uric acid, putrescine and primary n-alkylamines as the sole source of carbon, nitrogen and energy. Antonie Van Leeuwenhoek 1984; 50:369-78; PMID:6543110; http://dx.doi.org/10.1007/BF00394651
- Böer E, Breuer FS, Weniger M, Denter S, Piontek M, Kunze G. Large-scale production of tannase using the yeast Arxula adeninivorans. Appl Microbiol Biotechnol 2011; 92:105-14; PMID:21559827; http://dx.doi.org/10.1007/s00253-011-3320-5
- Sakai T, Jun HK. Purification and characterisation of adenine deaminase in Pseudomonas synxantha. J Ferment Technol 1978; 56:257-65.
- Cheng X, Yang B, Liu D, Chen G, Chen Y, Huang RF, Jiang YS. Genetic engineering of bacteria that can produce urate oxidase. Intern Med 2012; 2:4; http://dx.doi.org/10.4172/2165-8048.1000114

