Abstract
IBS is a prevalent functional gastrointestinal disorder, in which the microbiota has been demonstrated to play a role. An increasing number of studies have suggested how probiotics may alleviate IBS symptoms and several mechanisms of action have been proposed.
In the present study we characterized the intestinal microbiota of 19 subjects suffering from diagnosed IBS using a fully validated High Taxonomic Fingerprint Microbiota Array (HTF-Microbi.Array). We demonstrated that the IBS microbiota is different from that of healthy individuals due to an unbalance in a number of commensal species, with an increase in relative abundance of lactobacilli, B. cereus and B. clausii, bifidobacteria, Clostridium cluster IX and E. rectale, and a decrease in abundance of Bacteroides/Prevotella group and Veillonella genus. Additionally, we demonstrated that some bacterial groups of the human intestinal microbiota, recently defined as pathobionts, are increased in concentration in the IBS microbiota.
Furthermore, we aimed at investigating if the daily administration of a novel probiotic yogurt containing B. animalis subsp lactis Bb12 and K. marxianus B0399, recently demonstrated to have beneficial effects in the management of IBS symptoms, could impact on the biostructure of IBS microbiota, modulating its composition to counteract putative dysbiosis found in IBS subjects. Notably, we demonstrated that the beneficial effects associated to the probiotic preparation are not related to significant modifications in the composition of the human intestinal microbiota.
Introduction
Human beings are colonized by several microbial communities, which have the potential to impact on the host health. The vast majority of these bacterial populations thrive in the GIT and constitute the human intestinal microbiota,Citation1 whose main functions are related to nutrient processing, energy production, synthesis of cellular components and shaping of host innate and adaptive immunity, thus contributing to the maintenance of immune homeostasis in the gut.Citation2,Citation3 In physiological conditions, despite conservation at the highest taxonomic ranks, the intestinal microbiota is markedly individual-specific at species level, and a host-driven “top-down” assembly of the symbiotic microbial community has been suggested.Citation4 Recently, it has been hypothesized that high taxonomic level unbalances of the human gut microbiota can be responsible for important modifications of the host physiological status, being associated with a number of gastrointestinal disorders, i.e., IBS, IBD and colorectal cancer, as well as obesity and type 2 diabetes, cardiovascular disease and atopic syndrome.Citation5-Citation7
IBS is the prevalent functional GIT disorder with a worldwide prevalence of 10–20%.Citation8 IBS sufferers can be grouped into three main symptom subtypes: diarrhea-predominant IBS (D-IBS), constipation-predominant IBS (C-IBS) and mixed bowel habit IBS (M-IBS). The cause of the disease is thought to be multifactorial and may include dysmotility, abnormal gut sensation, genetic, microbial and dietary factors, as well as low-grade inflammation.Citation9 Several studies using qPCR, FISH and sequencing of 16S rDNA libraries reported an intestinal dysbiosis in patients suffering from IBS, in terms of specific compositional changes associated with the disorder.Citation9-Citation13 However, in most of these studies, the overall microbiota was not covered, as the quantified bacteria were predetermined according to primer or probe sequences. Only very recently, Rajilic-Stojanovic et al.Citation14 performed an in depth analysis of the human intestinal microbiota in IBS using a phylogenetic microarray targeting the bacterial 16S rRNA gene, demonstrating a significant decrease of Bacteroidetes (mainly belonging to Bacteroides and Prevotella), bifidobacteria and fecalibacterium prausnitzii, and an increase in Firmicutes.
In the last decade, the therapeutic role of probiotics in the IBS management has been proposed and different studies supported the efficacy of probiotics in alleviating IBS.Citation15,Citation16
In the present study, we aimed at characterizing the intestinal microbiota of 19 subjects suffering from diagnosed IBS (10 D-IBS, 5 M-IBS, 4 C-IBS), enrolled in a monocentric trial, and evaluating the impact of a novel probiotic yogurt on their intestinal microbiota.
The novel dairy probiotic preparation investigated in the present study contained Lactobacillus delbrueckii subsp bulgaricus and Streptococcus thermophilus, and was supplemented with Bifidobacterium animalis subsp lactis Bb12, a probiotic strain previously described as useful to manage IBS,Citation17,Citation18 and Kluyveromyces marxianus B0399, a novel probiotic lactic yeast recently characterized for its potentially beneficial properties.Citation19
A fully validated High Taxonomic Fingerprint Microbiota Array (HTF-Microbi.Array) was used to characterize the intestinal microbiota of the IBS subjects, before and after the probiotic intervention.Citation20 This DNA microarray, based on the LDR technology,Citation21 is a highly specific, reproducible and sensitive tool that enables specific detection and approximate relative quantification of 16S rRNAs from 30 phylogenetically related groups of the human intestinal microbiota. Differently from other DNA microarray platform already reported in literature, the HTF-Microbi.array, which allows to detect and quantify about the 95% of the human intestinal microbiota,Citation22 is specifically designed to monitor the high level taxonomic unbalances of the core functional microbiome that have an impact on the host physiological state.Citation5,Citation6,Citation23 Conversely, it remains blind to the species-level inter-individual variability.
To assess the most relevant unbalances characterizing the IBS microbiota, we compared the compositional data of the fecal microbiota of the IBS subjects recruited in this study with those deriving from the analysis by HTF-Microbi.Array of a cohort of 24 healthy adults, obtained in previous descriptive studies (Candela et al., 2010;Citation20 Candela et al., 2011, personal communication). Furthermore, since the dairy probiotic tested in this study has been previously demonstrated, in a monocentric human trial, to provoke an improvement of bloating, bowel movement abnormality, as well as reduction in abdominal pain in IBS patients,Citation24 we evaluated if the IBS-associated unbalances of the intestinal microbiota demonstrated in this study were reverted by the probiotic yogurt.
Results
Characterization of the intestinal microbiota in IBS subjects: Comparison with healthy subjects and impact of the probiotic intervention
High taxonomic fingerprints of the fecal microbiota of the IBS subjects were depicted by HTF-Microbi.Array, and compared with those of healthy subjects deriving from previous descriptive studies (Candela et al., 2010;Citation20 Candela et al., 2011, personal communication) (File S1).
The main bacterial groups of the IBS microbiota were Clostridium cluster IV and XIV (25% and 21% of the total microbiota, respectively), followed by Bacteroides/Prevotella (9.1%). Other subdominant bacterial groups found at relevant concentration in the IBS microbiota were lactic acid bacteria (7.8%, summing the hits of Lactobacillaceae family and those of the Lactobacillus species targeted by the HTF-Microbi.Array), as well as Veillonella genus (5.7%), Bacilli class (2.9%) and Bifidobacteriaceae family (1.2%).
Multivariate redundancy analysis of the relative abundance of targeted bacterial groups/species highlighted that the microbiota of IBS subjects is significantly different from that of healthy individuals (p < 0.05). Triplot of the RDA of the composition of the fecal microbiota of healthy and IBS subjects demonstrated that samples clearly separated on the basis of the health status (). Furthermore, D-IBS group was clearly separated from a cluster composed by samples belonging to M-IBS and C-IBS groups, which were more similar for their intestinal microbiota profiles.
Figure 1. Triplot of the RDA of the microbiota composition of subjects suffering from IBS and healthy individuals. Healthy subjects (HS), M-IBS patients (M), C-IBS patients (C) and D-IBS patients (D) are indicated by yellow rectangles, green diamonds, black circles and purple square, respectively. Constrained explanatory variables (HS, M, C, D) are indicated by filled red triangles. Black arrows indicate responding bacterial subgroups that explain more than 15% of the variability of the samples. First and second ordination axes are plotted, showing 12.3% and 5.4% of the total variability in the data set, respectively.
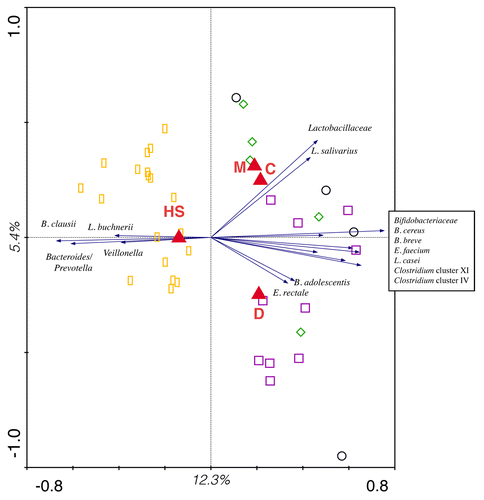
shows the bacterial groups significantly (p < 0.005) altered in IBS subjects with respect to healthy controls. In particular, the fecal microbiota of IBS subjects was demonstrated to be enriched in bacilli, Bifidobacteriaceae, Clostridium cluster IX, E. rectale and Lactobacillaceae. Notably, members of Enterobacteriaceae, E. faecium, C. difficile and Campylobacter spp were also demonstrated to be enriched in the IBS microbiota, with respect to the fecal microbiota of healthy subjects. Conversely, the IBS microbiota was depleted in concentration of Bacteroides/Prevotella group and Veillonella genus.
Table 1. Bacterial groups significantly altered in IBS subjects (IBS; Constipation IBS, C-IBS; Diarrhea IBS, D-IBS; Mixed IBS, M-IBS), with respect to healthy subjects (HS)
A number of significant variations in specific phylogenetic groups have been demonstrated not only between IBS and healthy controls, but also among the different IBS subtypes and healthy individuals ().
The comparison of the IBS microbiota composition before and after intake of the probiotic yogurt supplemented with B. animalis subsp lactis Bb12 and K. marxianus B0399 was performed. The microarray data sets of the fecal microbiota of the IBS subjects analyzed in the present study were hierarchically clustered on the basis of the signal intensity of the HTF-Microbi.Array oligonucleotide probes (). According to the main phylogenetic features of the fecal microbiota, two groupings were assessed. A marked inter-individual diversity was demonstrated, and the majority of the samples before and after intervention clustered together (12 out of 19 subjects). Therefore, no grouping according to the probiotic intervention was depicted.
Figure 2. Hierarchical clustering of the HTF-Microbi.Array profiles of IBS subjects before and after the probiotic administration. Microarray fingerprints at the baseline are indicated by t0, whereas fingerprints after the probiotic intervention are indicated by t1. Color intensity represents the relative bacterial abundance in the sample, in relation to the study population. Subjects whose t0 and t1 samples are not clustering together are marked with dots of different colors. Subjects whose t0 and t1 samples are clustering together are connected by line. Euclidean distance and Ward's clustering method were applied to log-transformed data.
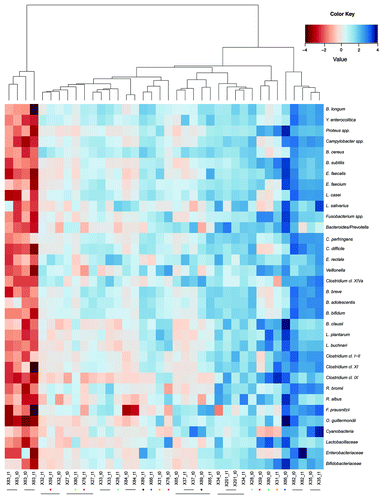
Differences in relative abundance of one or few bacterial groups constituting the human intestinal microbiota along the intervention were found in 7 out of 19 subjects, leading their samples (X28, X31, X59, X66, X78, X80, X89) not to cluster together at t0 and t1. However, these differences were accountable at the individual level and not shared among the 7 subjects. Therefore, the modifications of the microbiota profiles found in these 7 subjects between t0 and t1 did not impact on the relative abundance of the targeted microbial groups within the overall cohort ().
Moreover, PCR-DGGE analysis was used to retrieve an additional picture on the dynamics of the bacterial community before and after intervention. PCR-DGGE confirmed that the biodiversity of the intestinal microbiota was not influenced by the probiotic treatment, as assessed by the richness and Shannon indices (p > 0.05). Mean values of the richness index ranged from 17.5 (T0) to 19.6 (T1), whereas mean values of the Shannon index ranged from 2.75 (T0) to 3.02 (T1). Finally, the peak heights of DGGE densitometric curves were analyzed using the Mann-Whitney U-test, in order to assess if the most relevant single-species abundances were affected by the probiotic administration. No significant changes in species abundance were found when comparing T0 and T1.
Evaluation of the survival of K. marxianus B0399 along the probiotic intervention
The survival of K. marxianus B0399 along the probiotic intervention was tested using selective plate counting for lactic yeasts and semi-quantitative PCR-DGGE analysis followed by band identification.
Total count of fecal lactic yeasts showed negligible levels (< 100 CFU g−1 of faeces) at T0 in 15/19 subjects (75%), while the remaining 4 subjects had a basal concentration of (3.2 ± 0.6) × 103 CFU/g. Following the probiotic treatment, 16/19 subjects (84% of the study population, p < 0.001) were positive for yeast colonization, which reached a T1 concentration of (4.3 ± 1.2) × 105 CFUg-1. The presence of K. marxianus within the micro-eukaryotic fecal microbiota was confirmed by PCR-DGGE. PCR-DGGE analysis, whose sensitivity (~105 yeast cell mL−1) was not sufficient to detect K. marxianus at T0, confirmed the presence of a clear band corresponding to K. marxianus (99% sequence identity with K. marxianus 13MCHS 26S rRNA gene, File S2) at T1 in 14/19 subject (74% of the study population, p < 0.001).
Discussion
Recently, a number of studies investigated the unbalances that characterize the intestinal microbiota of patients suffering from IBS.Citation9-Citation14,Citation25 However, the rationale beyond differences in the microbiota composition between IBS patients and healthy individuals is still evolving. Furthermore, a growing number of studies have evaluated the response of IBS to probiotics, and few recent systematic reviews and meta-analyses suggested that probiotics appear to be, to varying extent, effective or at least promising in the amelioration of the well-being status of IBS subjects.Citation15,Citation16,Citation26
In the present study, we analzyed the fecal samples of 19 subjects suffering from IBS enrolled in a clinical trial for the evaluation of the efficacy of a new probiotic yogurt containing K. marxianus B0399 and B. animalis subsp lactis Bb12.Citation24 First, we evaluated the shift in the microbiota composition of the IBS patients before the probiotic administration, by comparing their microbiota profiles with those of a cohort of 24 healthy subjects, matched for sex and age, and previously characterized (Candela et al., 2010;Citation20 Candela et al., 2011, personal communication). Successively, we assessed the impact of the probiotic yogurt on the gut microbiota composition.
We demonstrated that IBS microbiota is different from that of healthy individuals due to an unbalance in a number of commensal species, with an increase in relative abundance of lactobacilli, B. cereus and B. clausii, bifidobacteria, Clostridium cluster IX and E. rectale, and a decrease in abundance of Bacteroides/Prevotella group and Veillonella genus. Furthermore, we demonstrated that some bacterial groups of the human intestinal microbiota, recently defined as pathobionts, are increased in concentration in the IBS microbiota. The so-called pathobionts are bacteria that can asymptomatically colonize the human GIT, but possessing pro-inflammatory characteristics they might have a role in causing disease when, due to a dysbiosis, they increase in concentration.Citation27 In the present study, members of the Enterobacteriaceae family, E. faecium, C. difficile and Campylobacter spp were demonstrated to be enriched in the IBS microbiota, with respect to the fecal microbiota of healthy subjects.
That the intestinal microbiota of subjects suffering from IBS deviates from the definition of a standard core microbiota in healthy conditions is a matter of fact, since an increasing number of studies evidenced peculiar modifications in the composition of the human intestinal microbial ecosystem correlated with health and disease status.Citation28,Citation29 However, the recent advances in the understanding of complex biological data deriving from metagenomics, metatranscriptomics and metabolomics approaches, are challenging the definitions of both standard core microbiota and IBS microbiota. In fact, these “-omics” techniques are giving novel insights on the dynamic interplay of the microbial species thriving human gut. To date, a limited number of phylum- and group-level differences have been demonstrated comparing IBS patients to healthy subjects, while several alterations in abundance at genus and species level have been identified, leading to results that are sometimes controversial.Citation25
In particular, our results are in accordance with those demonstrating an increase in abundance of Lactobacillus genus in IBS,Citation11,Citation13 which has been associated with augmented concentration of the organic acids propionate and acetate, that in turn were correlated with abdominal pain, bloating and anxiety by Tana et al.Citation13 Conversely, other studies indicated a depletion of lactobacilli as a characteristic of the IBS microbiota.Citation10,Citation12 Interestingly, a recent study performed using a DNA phylogenetic microarray by Rajilic-Stojanovic et al.Citation14 demonstrated a trend similar to that reported in this study, in relation to the dynamics of the Bacteroides/Prevotella group and bacilli, with the first group depleted in subjects suffering from IBS and the second one increased in abundance in the IBS microbiota. We also demonstrated that the IBS microbiota showed enrichment in bifidobacterial concentration, result which is in contrast with previous findings reporting decreased bifidobacterial concentrations in IBS patients.Citation12,Citation14 The HTF-Microbi.Array used in this study targets the entire Bifidobacteriaceae family and the B. longum, B. adolescentis, B. breve and B. bifidum species.
Numerous human studies and clinical trials have investigated the therapeutic benefit of probiotics in alleviating the symptoms of IBS, with a wide range of formulations and microbial species tested. Commonly used probiotic strains belong to Bifidobacterium or Lactobacillus genera, while less frequently used are strains of Propionibacterium freudenreichii, bacilli or yeasts.Citation30 Recently, we demonstrated in an in vitro study that K. marxianus B0399 possesses a number of beneficial properties, i.e., modulation of the immune response of PBMC and Caco-2 cells, impact on the metabolic activity of the intestinal microbiota and survival to simulated gastrointestinal environment, supporting its application as a probiotic.Citation19 The efficacy of a probiotic yogurt including K. marxianus B0399 and B. animalis subsp lactis Bb12 in the management of IBS has been investigated in an in vivo study.Citation20 The authors showed that these probiotics provoked an improvement in abdominal pain, bloating and bowel movement abnormality.
We characterized the intestinal microbiota of the 19 IBS patients enrolled in the above mentioned clinical study, with the aim of evaluating the impact of the probiotic administration on the IBS-associated unbalances of the intestinal microbiota.
Using both HTF-Microbi.Array and PCR-DGGE, we demonstrated that the supplementation of K. marxianus B0399 and B. animalis subsp lactis Bb12 for 4 weeks did not modulate the composition of the microbiota in the IBS patients. Indeed, a marked inter-individual diversity was evident, since the majority of the samples before and after intervention clustered together, and no groupings according to the probiotic intervention were depicted. Similarly, Shannon and richness indices of DGGE gels were not modified by the 4-week probiotic administration. At the light of the most recent findings, our results are in agreement with an increasing number of studies demonstrating that probiotic administration is often not accompanied by compositional modulations of the intestinal microbiota in subjects suffering from IBS,Citation31 microbiota-mediated systemic disordersCitation32 and in healthy conditions.Citation33
In conclusion, we improved the knowledge about the peculiar modifications characterizing the intestinal microbiota of subjects suffering from IBS. Furthermore, we demonstrated that the beneficial effects of the probiotic yogurt containing K. marxianus B0399 and B. animalis subsp lactis Bb12 are not associated to significant modifications of the human intestinal microbiota. These results open a new scenario about the necessity of characterizing the mechanism of action of clinically relevant probiotic strains not only toward the composition of the gut microbiota, but also taking into account its functionality.
Materials and Methods
Subjects and study design
The study group consisted of 19 subjects suffering from diagnosed IBS, who were enrolled in the intervention study. IBS patients (mean age = 33.6 ± 9.1) fulfilled the Rome III criteria for the diagnosis of IBS.Citation8 Exclusion criteria included pregnancy or lactation, chronic intestinal disease (i.e., inflammatory bowel disease or celiac disease) or severe systemic disorders, lactose intolerance or food allergies. Patients who in the 2 mo prior to study entry had taken medication, such as antibiotics, corticosteroids or functional foods containing pre- or probiotics, were also excluded from the study. Each subject signed the informed consent prior to enter the study. The study protocol was conforming to the ethical guidelines of the “World Medical Association Declaration of Helsinki.”
IBS patients were subjected to a 4-week study period, and were daily receiving a probiotic yogurt containing L. delbrueckii subsp bulgaricus and S. thermophilus and supplemented with K. marxianus B0399 and B. animalis subsp lactis Bb12. The total daily amount of K. marxianus B0399 was 1–4 × 107 CFU, while the amount of B. animalis subsp lactis Bb12 was 3–5 × 109 CFU. Consumption of other probiotics was not allowed during the intervention. All subjects were advised to follow their usual dietary habits and not to undertake any medication.
For the comparative analysis of the IBS microbiota composition before the probiotic intervention, a cohort of 24 healthy subjects comparable for age and sex and already characterized for their intestinal microbiota profiling was considered. These compositional data were retrieved using the same approaches, in relation to fecal sample collection and storage, sample processing and DNA microarray analysis, undertaken in this study (Candela et al., 2010;Citation20 Candela et al., 2011, personal communication).
Fecal samples
During the study, two fecal samples were collected from each subject suffering from IBS. Samples were taken the day before beginning the probiotic supplementation for assessing the individual baseline, and after the 4-week supplementation (not later than 24 h after the end of the probiotic intervention). Samples were immediately stored in anaerobic containers and frozen within 4 h to -70°C until analysis.
Extraction and purification of microbial DNA from fecal samples
Total microbial DNA was extracted using QIAamp DNA Stool Mini Kit (Qiagen) with a modified protocol, as previously described by Biagi et al.Citation34 Final DNA concentration was determined by using NanoDrop ND-1000 (NanoDrop Technologies).
Characterization of the intestinal microbiota by HTF-Microbi.Array
The intestinal microbiota of the IBS subjects enrolled in the study was characterized using the fully validated DNA microarray HTF-Microbi.Array, which target 30 phylogenetically related groups (File S3). The analysis was performed at the baseline and after the 4-week probiotic supplementation. 16S rRNA was amplified using universal forward primer 16S27F and reverse primer r1492, following the protocol previously described.Citation20,Citation21 PCR products were purified by using a Wizard SV gel and PCR clean-up System purification kit (Promega Italia), according to the manufacturer's instructions, eluted in 20 μl of sterile water, and quantified with the DNA 7500 LabChip Assay kit and BioAnalyzer 2100 (Agilent Technologies).
Ligase Detection Reaction and hybridization of the products on the universal arrays were performed according to the protocol described by Castiglioni et al.Citation21 except for the probe annealing temperature, set at 60°C.
PCR-DGGE analysis of the fecal samples
Studies on the microbial DNA fingerprints derived from PCR-DGGE analysis were performed for the IBS subjects, at the baseline and following the 4-week probiotic supplementation. Amplification of the V2-V3 region of the bacterial 16S rRNA gene was performed using the universal eubacterial primers GCclamp-HDA1 and HDA2, according to the protocol previously described by Maccaferri et al.Citation35 DGGE gel images were analyzed using the FPQuest Software Version 4.5 (Bio-Rad). In order to compensate for gel-to-gel differences and external distortion to electrophoresis, the DGGE patterns were aligned and normalized using an external reference ladder composed by known bacterial species. After normalization, bands were defined for each sample using the appropriate densitometric curves. Bands constituting less than 1% of the total band area were omitted from further analysis. Similarity between DGGE profiles was determined by calculating the Pearson correlation. Clustering of the sample profiles was done using the UPGMA algorithm. Additionally, a Shannon diversity index was calculated to investigate the structural diversity of the microbial community.Citation36
Culture-independent and -dependent detection of K. marxianus B0399 in the fecal microbiota
Dynamics of yeast population and detection of the administered yeast during the study were assessed by PCR-DGGE and selective plate counting, respectively. Approximately 250 nucleotides of the 5′-end region of the 26S rRNA gene were amplified by PCR using the yeast-universal primer set GC-clamped NL1 and LS2, according to Cocolin et al.Citation37 The PCR-DGGE experimental protocol was slightly modified by performing annealing at 56°C for 25 sec and extension at 72°C for 30 sec, in order to prevent cross-amplification of bacterial DNA. Band identity was confirmed by comparison of the positions in the gel length with those of reference yeast DNA.
Detection of the survival and quantification of the growth of K. marxianus B0399 along the intervention were performed by plate counting at 37°C in lactic-yeast selective MV2 agar (lactose, 40 g L−1; casein, 20 g L−1; peptone, 7.5 g L−1; yeast extract, 1.5 g L−1; agar 10% w/v). 1:5 (w/w) fecal dilutions in anaerobic PBS [0.1 mol L−1 PBS (pH 7.4)] were prepared and the samples were homogenized in a stomacher (Seward Ltd.) for 2 min. After homogenization, fecal samples were serially diluted using 10-fold serial dilutions down to a final dilution of 10−5. All plating counts were performed in triplicate.
Statistics
All arrays were scanned with ScanArray 5000 scanner (Perkin Elmer Life Sciences), at 10 μm resolution. Fluorescent images were obtained with different acquisition parameters for both laser power and photo-multiplier gain, in order to avoid saturation. Fluorescence intensities were quantitated by ScanArray Express 3.0 software, using the “Adaptive circle” option, letting diameters vary from 60 to 300 μm. To assess whether a probe pair was significantly above the background (i.e., was present or not), one-sided t-test was performed. Nonparametric Kruskal-Wallis test, followed by Bonferroni correction for multiple comparisons, was used to determine the statistical differences among the IBS subtypes and/or treatment conditions. A p < 0.05 was considered statistically significant. Hierarchical clustering of HTF-Microbi.Array profiles was performed using the statistical software R (http://www.r-project.org). The Euclidean distance among sample profiles was calculated and Ward's method was used for agglomeration. Redundancy analysis and RDA ordination diagram were performed using CANOCO for Windows 4.02 and CanoDraw 3.10 (Microcomputer Power), respectively. Monte Carlo permutation test was done at 199 random permutations, in order to assess significant differences.
Bacterial counts and DGGE parameters were analyzed by one-way ANOVA, using Tukey’s post-test analysis when the overall P value of the experiment was below the value of significance (p < 0.05). An additional paired t-test was applied in order to assess the significance of the results of single pairs of data. Analyses were performed using GraphPad Prism 5.0 (GraphPad Software).
| Abbreviations: | ||
| DGGE | = | denaturing gradient gel electrophoresis |
| GIT | = | gastro-intestinal tract |
| IBS | = | irritable bowel syndrome |
| IBD | = | inflammatory bowel diseases |
| LDR | = | ligase detection reaction |
| RDA | = | redundancy analysis |
| UPGMA | = | unweighted pair group method using arithmetic average |
Additional material
Download Zip (173.4 KB)Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature 2007; 449:804 - 10; http://dx.doi.org/10.1038/nature06244; PMID: 17943116
- Gosalbes MJ, Durbán A, Pignatelli M, Abellan JJ, Jiménez-Hernández N, Pérez-Cobas AE, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One 2011; 6:e17447; http://dx.doi.org/10.1371/journal.pone.0017447; PMID: 21408168
- Jarchum I, Pamer EG. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunol 2011; 23:353 - 60; http://dx.doi.org/10.1016/j.coi.2011.03.001; PMID: 21466955
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A 2010; 107:18933 - 8; http://dx.doi.org/10.1073/pnas.1007028107; PMID: 20937875
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008; 453:620 - 5; http://dx.doi.org/10.1038/nature07008; PMID: 18509436
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature 2009; 457:480 - 4; http://dx.doi.org/10.1038/nature07540; PMID: 19043404
- Maccaferri S, Biagi E, Brigidi P. Metagenomics: key to human gut microbiota. Dig Dis 2011; 29:525 - 30; http://dx.doi.org/10.1159/000332966; PMID: 22179207
- Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology 2006; 130:1480 - 91; http://dx.doi.org/10.1053/j.gastro.2005.11.061; PMID: 16678561
- Codling C, O’Mahony L, Shanahan F, Quigley EM, Marchesi JR. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci 2010; 55:392 - 7; http://dx.doi.org/10.1007/s10620-009-0934-x; PMID: 19693670
- Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol 2005; 100:373 - 82; http://dx.doi.org/10.1111/j.1572-0241.2005.40312.x; PMID: 15667495
- Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Paulin L, Corander J, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 2007; 133:24 - 33; http://dx.doi.org/10.1053/j.gastro.2007.04.005; PMID: 17631127
- Kerckhoffs AP, Samsom M, van der Rest ME, de Vogel J, Knol J, Ben-Amor K, et al. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol 2009; 15:2887 - 92; http://dx.doi.org/10.3748/wjg.15.2887; PMID: 19533811
- Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M, Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil 2010; 22:512 - 9, e114-5; PMID: 19903265
- Rajilić-Stojanović M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011; 141:1792 - 801; http://dx.doi.org/10.1053/j.gastro.2011.07.043; PMID: 21820992
- Marteau P. Probiotics in functional intestinal disorders and IBS: proof of action and dissecting the multiple mechanisms. Gut 2010; 59:285 - 6; http://dx.doi.org/10.1136/gut.2008.173690; PMID: 20207630
- Spiller R. Review article: probiotics and prebiotics in irritable bowel syndrome. Aliment Pharmacol Ther 2008; 28:385 - 96; http://dx.doi.org/10.1111/j.1365-2036.2008.03750.x; PMID: 18532993
- Simrén M, Ohman L, Olsson J, Svensson U, Ohlson K, Posserud I, et al. Clinical trial: the effects of a fermented milk containing three probiotic bacteria in patients with irritable bowel syndrome - a randomized, double-blind, controlled study. Aliment Pharmacol Ther 2010; 31:218 - 27; PMID: 19863495
- Søndergaard B, Olsson J, Ohlson K, Svensson U, Bytzer P, Ekesbo R. Effects of probiotic fermented milk on symptoms and intestinal flora in patients with irritable bowel syndrome: a randomized, placebo-controlled trial. Scand J Gastroenterol 2011; 46:663 - 72; http://dx.doi.org/10.3109/00365521.2011.565066; PMID: 21443416
- Maccaferri S, Klinder A, Brigidi P, Cavina P, Costabile A. Potential probiotic Kluyveromyces marxianus B0399 modulates the immune response in Caco-2 cells and peripheral blood mononuclear cells and impacts the human gut microbiota in an in vitro colonic model system. Appl Environ Microbiol 2012; 78:956 - 64; http://dx.doi.org/10.1128/AEM.06385-11; PMID: 22156412
- Candela M, Consolandi C, Severgnini M, Biagi E, Castiglioni B, Vitali B, et al. High taxonomic level fingerprint of the human intestinal microbiota by ligase detection reaction--universal array approach. BMC Microbiol 2010; 10:116; http://dx.doi.org/10.1186/1471-2180-10-116; PMID: 20398430
- Castiglioni B, Rizzi E, Frosini A, Sivonen K, Rajaniemi P, Rantala A, et al. Development of a universal microarray based on the ligation detection reaction and 16S rrna gene polymorphism to target diversity of cyanobacteria. Appl Environ Microbiol 2004; 70:7161 - 72; http://dx.doi.org/10.1128/AEM.70.12.7161-7172.2004; PMID: 15574913
- Rajilić-Stojanović M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 2007; 9:2125 - 36; http://dx.doi.org/10.1111/j.1462-2920.2007.01369.x; PMID: 17686012
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res 2009; 19:1141 - 52; http://dx.doi.org/10.1101/gr.085464.108; PMID: 19383763
- Lisotti A, Cornia GL, Morselli-Labate AM, Sartini A, Turco L, Grasso V, et al. Effects of a fermented milk containing Kluyveromyces marxianus B0399 and Bifidobacterium lactis Bb12 in patients with irritable bowel syndrome. Minerva Gastroenterol Dietol 2011; 57 (Suppl. 1 n.2):1-12.
- Salonen A, de Vos WM, Palva A. Gastrointestinal microbiota in irritable bowel syndrome: present state and perspectives. Microbiology 2010; 156:3205 - 15; http://dx.doi.org/10.1099/mic.0.043257-0; PMID: 20705664
- Ringel Y, Ringel-Kulka T. The rationale and clinical effectiveness of probiotics in irritable bowel syndrome. J Clin Gastroenterol 2011; 45:Suppl S145 - 8; http://dx.doi.org/10.1097/MCG.0b013e31822d32d3; PMID: 21992954
- Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes?. Nat Rev Immunol 2010; 10:735 - 44; http://dx.doi.org/10.1038/nri2850; PMID: 20865020
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev 2010; 90:859 - 904; http://dx.doi.org/10.1152/physrev.00045.2009; PMID: 20664075
- Wallace TC, Guarner F, Madsen K, Cabana MD, Gibson G, Hentges E, et al. Human gut microbiota and its relationship to health and disease. Nutr Rev 2011; 69:392 - 403; http://dx.doi.org/10.1111/j.1753-4887.2011.00402.x; PMID: 21729093
- Wassenaar TM, Klein G. Safety aspects and implications of regulation of probiotic bacteria in food and food supplements. J Food Prot 2008; 71:1734 - 41; PMID: 18724773
- Michail S, Kenche H. Gut microbiota is not modified by randomized, double-blind, placebo-controlled trial of VSL#3 in diarrhea-predominant Irritable Bowel Syndrome. Probiotics Antimicrob Proteins 2011; 3:1 - 7; http://dx.doi.org/10.1007/s12602-010-9059-y; PMID: 22247743
- Larsen N, Vogensen FK, Gøbel R, Michaelsen KF, Abu Al-Soud W, Sørensen SJ, et al. Predominant genera of fecal microbiota in children with atopic dermatitis are not altered by intake of probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium animalis subsp. lactis Bi-07. FEMS Microbiol Ecol 2011; 75:482 - 96; http://dx.doi.org/10.1111/j.1574-6941.2010.01024.x; PMID: 21204871
- Vitali B, Ndagijimana M, Cruciani F, Carnevali P, Candela M, Guerzoni ME, et al. Impact of a synbiotic food on the gut microbial ecology and metabolic profiles. BMC Microbiol 2010; 10:4; http://dx.doi.org/10.1186/1471-2180-10-4; PMID: 20055983
- Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 2010; 5:e10667; http://dx.doi.org/10.1371/journal.pone.0010667; PMID: 20498852
- Maccaferri S, Vitali B, Klinder A, Kolida S, Ndagijimana M, Laghi L, et al. Rifaximin modulates the colonic microbiota of patients with Crohn’s disease: an in vitro approach using a continuous culture colonic model system. J Antimicrob Chemother 2010; 65:2556 - 65; http://dx.doi.org/10.1093/jac/dkq345; PMID: 20852272
- Magurran A. Measuring biological diversity. Oxford: Blackwell Publishing; 2004.
- Cocolin L, Bisson LF, Mills DA. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol Lett 2000; 189:81 - 7; http://dx.doi.org/10.1111/j.1574-6968.2000.tb09210.x; PMID: 10913870
