Abstract
The fungus Candida albicans and the gram-positive bacterium Enterococcus faecalis are both normal residents of the human gut microbiome and cause opportunistic disseminated infections in immunocompromised individuals. Using a nematode infection model, we recently showed that co-infection resulted in less pathology and less mortality than infection with either species alone and this was partly explained by an interkingdom signaling event in which a bacterial-derived product inhibits hyphal morphogenesis of C. albicans. In this addendum we discuss these findings in the contest of other described bacterial-fungal interactions and recent data suggesting a potentially synergistic relationship between these two species in the mouse gut as well. We suggest that E. faecalis and C. albicans promote a mutually beneficial association with the host, in effect choosing a commensal lifestyle over a pathogenic one.
Keywords: :
Polymicrobial Infections: An Understudied Clinical Problem
There has been a general awareness that polymicrobial infections—those in which more than one species are identified from clinical samples—are associated with worse clinical outcomes than monomicrobial infections for decades, though large-scale epidemiological studies are lacking.Citation1-Citation3 Our laboratories study two pathogens that are commonly found together in polymicrobial infections, the fungus Candida albicans and the gram-positive bacterium Enterococcus faecalis. These two organisms trail only staphylococci in terms of prevalence in nosocomial sepsis and are frequently found together in clinical samples. While reports describing the co-isolation of these two species are common (see, for instance, refs. Citation4–Citation6), one large-scale retrospective analysis of clinical samples illustrated the propensity of these species to be found together. Hermann, et al.Citation7 separated 68,000 specimens from a two-year period at a large German teaching hospital into those with detectable Candida (~5,200) and those without, including samples from both sterile (blood, CSF) and non-sterile (feces, sputum, skin) sites. Overall, E. faecalis was twice as likely to be found in Candida-positive samples than Candida negative ones; enrichment of E. faecalis in the fungal samples was even more dramatic in blood samples and in patients in the ICU.Citation7
The frequency with which the two species are cultured from the same samples may reflect their similar biology within the host, may be due to synergistic interactions, or some combination of the two. Both species commonly colonize the gastrointestinal and urogenital tracts of humans and are considered opportunistic pathogens, meaning that bloodstream infections are rare in the absence of an underlying immunological dysfunction. Whether they might specifically interact with each other to alter their commensal population size, propensity to disseminate, or inherent virulence properties had not been addressed, but were reasonable questions given the clinical significance of this specific polymicrobial infection.
A Nematode Co-Infection Model Reveals Surprising Interactions
The model invertebrate Caenorhabditis elegans has been successfully employed as a system to study both bacterial and fungal pathogenesis,Citation8 including for both C. albicans and E. faecalis.Citation9,Citation10 It offers numerous advantages in terms of ease of use, cost, ethical considerations, and host genetics; most importantly, it has identified factors known to be relevant to virulence in mammals including the FsrB quorum sensing system in E. faecalis and hyphal morphogenesis in C. albicans.Citation9-Citation11 This model has also been used to investigate polymicrobial infections between C. albicans and Acinetobacter baumanii.Citation12 These precedents encouraged us to examine the effects of co-infection by these two species in the worm model.Citation13
In the laboratory, C. elegans are normally fed on a lawn of non-pathogenic E. coli grown on the surface of an agar medium. To prepare C. elegans for pathogen exposure, the worms are first allowed to develop on E. coli to the fourth larval stage (L4), which is the stage just before adulthood. Then the animals are transferred onto a lawn of pathogen. Survival is usually assayed over time on the solid medium.Citation10 For infection with C. albicans, we followed a variation on this protocol, which involved moving the animals into liquid medium after a few hours of exposure to C. albicans on solid medium.Citation12,Citation13 For co-infection, we either exposed the animals to E. faecalis on solid medium before or after exposure to C. albicans.Citation13 As expected, worms exposed to either species individually died far more quickly than those grown on the normal E. coli food source. This effect was particularly dramatic with C. albicans, where we observed florid hyphal growth that eventually penetrated the worm’s cortex, as had been described.Citation9 Next, we examined animals co-infected with both pathogens. Using fluorescently tagged cells, we showed that both species co-existed in the worm gut (). To our surprise, worms lived substantially longer when infected with both species relative to either one individually. The most dramatic effect was observed when the animals were first exposed to E. faecalis followed by C. albicans; worm survival was not statistically different from feeding on E. coli.Citation13 The effect was not as dramatic when the order of exposure was reversed. Therefore it is possible that exposure to E. faecalis somehow primes the worm’s innate immune system to better withstand a later exposure to C. albicans.
Figure 1. A C. elegans infection model reveals attenuation of virulence in C. albicans-E. faecalis coinfections. (A-D) Fluorescence imaging shows the two species mixing in the worm and the absence of hyphal growth in the coinfection. Worms were infected with yCherry labeled-C. albicans alone (A and B) or coinfected with GFP-labeled E. faecalis (C and D) and imaged using DIC (A and C) or fluorescence (B and D). Panels (E-H) Ultrastructural analysis of nematode physiology. Worms were infected with C. albicans alone (E and F) or with both species (G and H) before processing for transmission electron microscopy. The disruption of the normal gut physiology in the C. albicans-infected worms relative to the coinfection is apparent. The scale bar in panel F is 1 µm and applied to (E-H).
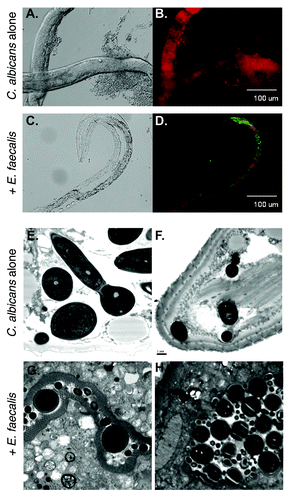
Another striking observation was that, no matter what the order of exposure, the presence of E. faecalis dramatically inhibited hyphal morphogenesis within the worm, reducing tissue damage. The intestinal pathologies seen with either pathogen alone were essentially absent in the co-infected animals (). Hyphal growth was also inhibited in mixed-species biofilms, but not in other strongly hyphal-inducing conditions in vitro, such as the presence of serum. The inhibition is mediated through a secreted compound under the influence of the FsrB system, as sterilized supernatants from wild-type but not fsrB- strains grown in media blocked morphogenesis and attenuated virulence of C. albicans in the worm model. Preliminary characterization indicated that the inhibitory compound is small and heat-stable. Its molecular identification is ongoing.Citation13 Notably, other bacterial species also secrete compounds that affect C. albicans biology. Hyphal-inducing, hyphal-inhibiting, and fungicidal activities have been observed, as will be summarized below.
However, this work leaves many questions beyond the nature of the inhibitory molecule. We cannot yet say how C. albicans attenuates E. faecalis killing in the worm or how the combination of the two species dramatically improves intestinal physiology. The applicability of these findings to virulence in a mammalian model has not been addressed either, though, as will be discussed below, there is some preliminary evidence in the literature of significant interactions between these two microbes in both a mouse infection model, and a mouse commensal model.
Under the Influence: Secreted Bacterial Signals that Affect C. albicans
It has long been known that microbes secrete products that can kill one another, a byproduct of competition for resources in their ecological niches. We have harnessed the occurrence of these compounds to our advantage, and most of our antimicrobial medicines are derived from naturally occurring microbial products. But microbes also produce secreted signals with more subtle effects than growth inhibition or death. Because C. albicans lives in the mammalian host in environments rich with prokaryotes, it is not surprising that this fungus responds to signals secreted by bacteria. Most studies have focused on hyphal morphogenesis, an easily observable phenotype. The first bacterium discovered that affects C. albicans morphogenesis was Pseudomonas aeruginosa. The secreted signal was identified as 3-oxo-C12 homoserine lactone (3C-HSL). HSLs are the primary molecules by which gram-negative species sense quorums, and it was notable that C. albicans sensed and responded to this signal, as inter-kingdom signaling was not well appreciated at the time.Citation14 The 3C-HSL resembles farnesol, a signaling molecule produced by C. albicans that inhibits hyphal formation through the Ras1-cAMP-Efg1 signaling pathway.Citation15,Citation16 Other gram-negatives such as Burkholderia cenocepacia and Xanthomonas campestris also inhibit C. albicans hyphal morphogenesis by producing decenoic acids that mimic the activity of farnesol.Citation17,Citation18 P. aeruginosa also releases toxic redox-active phenazines. At high concentrations, these compounds inhibit C. albicans growth, but recently, a new role has been discovered for phenazines at lower concentrations. They disfavor hyphal morphogenesis by inhibiting the respiratory chain; it is thought that the yeast form preferentially uses a fermentative mode of growth.Citation19
P. aeruginosa, A. baumannii and Salmonella typhimurium can each inhibit the Candida hyphal morphogenesis that occurs in the C. elegans model. Depending on the organism, this activity is totally or partially dependent on a secreted substance. Surprisingly, the compound does not appear to be 3C-HSL, as addition of pure 3C-HSL did not inhibit filamentation in C. albicans-infected worms. Also, loss of the primary A. baumannii HSL synthase, encoded by luxI, had no effect on its ability to inhibit Candida filamentation.Citation12 The inhibition by A. baumannii appeared to be partly dependent on the bacterium preferably attaching to the hyphae, killing this morphotype.Citation12S. typhimurium was also shown to inhibit hyphal morphogenesis in part by preferentially killing the hyphal form of C. albicans. In this case, the killing effect was not dependent on cell-to-cell contact and is due to an unidentified secreted substance found in stationary phase supernatants.Citation20
What about gram-positives other than E. faecalis? The staphylococci are not documented as affecting C. albicans hyphal morphogenesis, though there is evidence that they enhance biofilm formation when mixed with C. albicans.Citation12,Citation21 On the other hand, Streptococcus mutans, an oral microbe and the major etiological agent of cavities, was found to inhibit C. albicans hyphal morphogenesis in mixed species biofilmsCitation22 and this effect is mediated via a secreted competence-stimulating peptide (CSP),Citation23 part of a family of small and often modified peptides commonly used for quorum sensing in gram-positive species. Not surprisingly, several other streptococcal species that generate this peptide were also able to inhibit Candida morphogenesis, including S. gordonii, S. oralis, S. salivarius, and S. sobrinus. How this peptide exerts its effects on Candida remains mysterious.Citation23 Another report showed that a different S. gordonii strain actually enhanced C. albicans filamentation within biofilms in a mechanism somehow dependent on LuxS, which produces autoinducer-2.24
In summary, secreted bacterial products from a variety of species can inhibit or promote C. albicans filamentation. In many cases, particularly with Pseudomonas, the evidence points to a direct antagonism between the species in vitro. Yet, in vivo, the picture appears more complex. Pseudomonas ventilator-associated pneumonia is both more likely and more severe if the patient is colonized by Candida.Citation25 The presence of Pseudomonas exacerbates tissue damage in a model of Candida infection of burn wounds.Citation26 Biofilms formed between C. albicans and either S. aureus or S. gordonii are larger and more drug resistant than their monomicrobial counterparts.Citation24,Citation27 Clearly there are context-dependent interactions that prevent simple conclusions about the in vivo effect of mixed infections. The nematode infection model is a step between in vitro assays and the more challenging mammalian models.
E. faecalis-C. albicans Interactions in the Mammalian Host
The data reported in Cruz, et al.,Citation13 convincingly demonstrates that a co-infection of C. albicans with E. faecalis reduces tissue damage and extends lifespan of the worms relative to either monomicrobial infection. The presence of E. faecalis also dramatically lessens hyphal morphogenesis of C. albicans, a key virulence trait of this species.Citation28,Citation29 Moreover, this effect requires the E. faecalis Fsr two-component system, a central regulator of virulence in mammalian models.Citation30-Citation32 Thus, the phenomenon we have observed is closely associated with known virulence processes in both species.
Viewed through the traditional lens of microbial pathogenesis, our data showed a significant attenuation of virulence in the co-infection compared with the mono-infections. Yet, neither species tend to cause serious infections in healthy individuals, with the primary at-risk population being hospitalized and/or immunocompromised patients. Rather, in healthy people, both are primarily associated with the normal microbiota at multiple body sites, especially the gastrointestinal tract. So are the effects of this interkingdom co-infection really an attenuation of virulence? Perhaps C. albicans and E. faecalis have synergistic effects on colonization; that is, this interaction promotes a stable non-pathogenic association with the host.
Is there any evidence to support this idea? Yes, in fact, in striking work from Gary Huffnagle and colleagues at the University of Michigan, who have long been interested in the physiological effects of perturbing the gastrointestinal microbiota through the introduction of fungi. The mouse is an ideal model for this work because common lab strains do not harbor C. albicans as a commensal (some other Candida species can occasionally be isolated), allowing investigators to experimentally manipulate the fungal component of the microbiota. Noverr and Huffnagle showed some years ago that the introduction of C. albicans to the gut of uncolonized mice had demonstrable effects on systemic immune responses.Citation33 Numerous studies have addressed the disruption of gut bacterial populations by antibiotic treatment, and the subsequent recovery after cessation of the drug. These alterations are complex, dependent on the antibiotic and the individual, but the bacterial microbiota generally recovers with some loss of diversity (for a more detailed discussion, see ref. Citation34). Taking this one step further, Huffnagle asked whether the presence of C. albicans would alter the dynamics of bacterial recovery from antibiotic pressure.Citation35 Mice were given the broad-spectrum antibiotic cefoperazone for one week, at which point one group of animals were inoculated with C. albicans via intragastric gavage. The antibiotic was subsequently removed from both the inoculated and control groups and bacterial colonization in the stomach was monitored seven and 21 d later. While there were substantial differences in the overall population structure in both antibiotic-treated groups relative to untreated controls, the real surprise came when they focused on the lactic acid bacteria. In the no-Candida group, Enterococcus species were the first to recover, but were replaced by lactobacilli within three weeks. In stark contrast, the C. albicans-treated animals remained dominated by Enterococcus, in particular by E. faecalis, the species that synergized with C. albicans in our studies in the worm gut.Citation13,Citation35 A subsequent study focused on the cecum showed very similar results.Citation36 Notably, C. albicans-induced gastritis was much worse in gnotobiotic mice than it was in antibiotic-treated normal mice, indicating that even an impaired bacterial microbiota can suppress C. albicans-associated pathology.Citation35
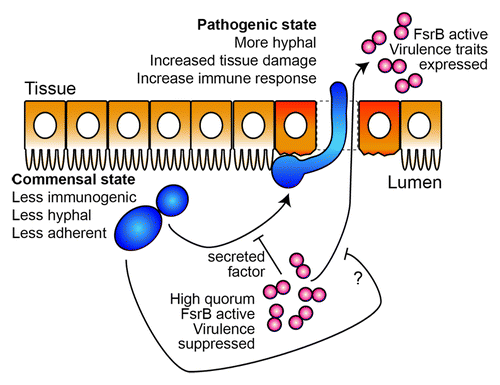
This work demonstrates that C. albicans promotes E. faecalis growth in the mammalian gastrointestinal tract, though it has not been determined whether this is through fungal antagonism of lactobacilli or synergism with enterococci. The latter would be consistent with our studies in C. elegans. presents a highly speculative model whereby interspecies interactions inhibit elaboration of virulence traits to promote a commensal interaction with the host. This working model serves as a basis for our future investigations and this model is complicated by the clinical picture, in which enterococci and Candida comprise polymicrobial infections associated with worse outcomes. Unfortunately, there is only one, decades-old, study in the literature that assesses the potential synergies between these two microbes in a mammalian infection model. Carlson infected mice by intraperitoneal injection with sub-letahal doses of C. albicans and E. faecalis.Citation37 Five days later, the animals were sacrificed and the blood, pancreas, kidney, spleen and peritoneum were assessed for CFUs of both E. faecalis and C. albicans. At doses of E. faecalis that do not normally cause detectable dissemination (107), the addition of C. albicans resulted in significant numbers of E. faecalis in all tissues tested. In fact, even with an inoculum as low as 103, E. faecalis was still detectable in some tissues. The synergism was not mutual; sublethal doses of the bacteria were not observed to stimulate C. albicans infectivity.Citation37 C. albicans enhancement of bacterial infection was not limited to E. faecalis, but also observed for Staphylococcus aureus and Serratia marcescens. A more recent study, examining the effects of C. albicans on S. aureus suggests that a heightened inflammatory response occurs when both microbes are injected together, which could account for the poorer outcome.Citation38
Figure 2. A speculative model for Candida-Enterococcus interactions in the gut. In the worm gut, C. albicans transitions from a commensal mode characterized by yeast-form growth to an invasive, hyphal and pathogenic state. This transition is inhibited by the presence of E. faecalis through molecule(s) secreted in an FsrB-dependent manner, thus maintaining the commensal association with the host. Likewise, mono-infection with E. faecalis imposes significant gut pathology, which is reduced in the presence of C. albicans through an unknown mechanism. In the mammalian gut, the work from Hufnagel and colleagues suggests that these two species also promote a commensal association with the host which we speculate can be disrupted by changes to microbe or host factors leading to infection. C. albicans cells are shown in blue, E. faecalis in red, and gut epithelial cells in orange.
The Complete Microbiota
Interactions among and between microbial kingdoms certainly play key roles in the structure of the human microbiota and its influence on health. An emerging therapy for Clostridium difficile infections, which are particularly refractory to antibiotic therapy, is the reconstitution of the microbiota via fecal transplants from healthy donors (rev. in ref. Citation39). An alternative remedy for vaginal candidiasis is the topical application of yogurt with active lactobacilli (see ref. Citation40). A very recent study demonstrated a dramatic difference in the penile microbiome before and after circumcision that may underlie the different susceptibilities to HIV and other sexually transmitted diseases.Citation41 There are and certainly will be additional examples of inter-microbial interactions that alter disease states or severity.
The move from culture-based to sequence-based determination of microbial populations has offered tremendous opportunities to understand the diversity, structure and dynamics of the microbiota. And, indeed, a vast array of literature has described the bacterial microbiome at various body sites, after specific iatrogenic interventions, and during development. The questions being asked are so diverse that a recent paper compared the microbiome of dog-owning families with their pets.Citation42
The specific and relevant interkingdom interactions we and others have identified between C. albicans and E. faecalis, P. aeruginosa and S. aureus, streptococci and others have come because Candida is a known component of the microbiota. Despite significant recent efforts to characterize the human-associated microbiome, our knowledge of the eukaryotic component remains reudimentary, because studies of the “complete” microbiome routinely focus on the bacterial-specific 16S rDNA, excluding eukaryotic microbes, both fungi and protozoans. This is fortunately changing thanks to the sequencing revolution: the landmark Human Microbiome Project, which characterized clinical specimens from 242 people, included some whole genome sequencing in which lower eukaryotes were identified, though most of the data were still based on 16S sequencing.Citation43,Citation44 A report describing the fungal microbiome of the skin appeared very recently, showing a community that differs both between individuals and between anatomical locations on the same person. Though dominated by species of the genus Malassezia, many fungal taxa were represented.Citation45 This is probably the most complete description of the fungal microbiome to date and hopefully these efforts will continue to increase our understanding of the diversity of the eukaryotic component.
What we do know of the eukaryotic microbiota hints at fascinating biology. C. albicans appears to have a molecular mechanism to suppress its own population size in the gut, for reasons unknown.Citation46 A limited study of the oral “mycobiome” from 20 individuals identified a total of 85 fungal genera, of which 15 were present in at least 20% of the subjects, with Candida as the most abundant.Citation47 Very limited 18S sequencing in a handful of people has suggested that the most abundant eukaryote in the human gut is not Candida, but the protozoan Blastocystis, with Galactomyces as the predominant fungal species.Citation48 Neither species has been extensively studied. Patients with hepatitis B or with inflammatory bowel disease have a more diverse fungal community than healthy controls.Citation49,Citation50 Most recently, a polymorphism in Dectin-1, a β-glucan receptor critical for antifungal recognition, has been associated with severity of ulcerative colitis.Citation51 How these organisms might affect the bacterial microbiome has simply not been addressed, but it is certainly reasonable to speculate that fungal burden and the composition of the microbiota contributes to the propensity to disseminate in at-risk patients.
In summary, our work has identified a specific interaction between C. albicans and E. faecalis that promotes the commensal form over the pathogenic, at least partly through suppression of fungal morphogenesis by a bacterially-derived molecule. This work is consistent with a potentially synergistic relationship between these two species uncovered by experimental manipulation of the murine gutCitation35,Citation36 and with recent studies of interactions between C. albicans and P. aeruginosa, S. aureus, and others (see ref. Citation52). We predict that many other interkingdom interactions will be identified with direct relevance to human health and a fascinating underlying biology.
Acknowledgments
Preparation of this manuscript was supported by Public Health Service Grants AI075091 to MCL and AI076406 to DAG.
Submitted
05/21/2013
Revised
06/30/2013
Accepted
08/04/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- Hermans PE, Washington JA 2nd. Polymicrobial bacteremia. Ann Intern Med 1970; 73:387 - 92; http://dx.doi.org/10.7326/0003-4819-73-3-387; PMID: 4917179
- McKenzie FE. Case mortality in polymicrobial bloodstream infections. J Clin Epidemiol 2006; 59:760 - 1; http://dx.doi.org/10.1016/j.jclinepi.2005.12.009; PMID: 16765282
- Rolston KV, Bodey GP, Safdar A. Polymicrobial infection in patients with cancer: an underappreciated and underreported entity. Clin Infect Dis 2007; 45:228 - 33; http://dx.doi.org/10.1086/518873; PMID: 17578784
- Gullberg RM, Homann SR, Phair JP. Enterococcal bacteremia: analysis of 75 episodes. Rev Infect Dis 1989; 11:74 - 85; http://dx.doi.org/10.1093/clinids/11.1.74; PMID: 2916096
- Klotz SA, Chasin BS, Powell B, Gaur NK, Lipke PN. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn Microbiol Infect Dis 2007; 59:401 - 6; http://dx.doi.org/10.1016/j.diagmicrobio.2007.07.001; PMID: 17888612
- Sutter D, Stagliano D, Braun L, Williams F, Arnold J, Ottolini M, Epstein J. Polymicrobial bloodstream infection in pediatric patients: risk factors, microbiology, and antimicrobial management. Pediatr Infect Dis J 2008; 27:400 - 5; http://dx.doi.org/10.1097/INF.0b013e31816591be; PMID: 18398386
- Hermann C, Hermann J, Munzel U, Rüchel R. Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 1999; 42:619 - 27; http://dx.doi.org/10.1046/j.1439-0507.1999.00519.x; PMID: 10680438
- Marsh EK, May RC. Caenorhabditis elegans, a model organism for investigating immunity. Appl Environ Microbiol 2012; 78:2075 - 81; http://dx.doi.org/10.1128/AEM.07486-11; PMID: 22286994
- Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog 2007; 3:e18; http://dx.doi.org/10.1371/journal.ppat.0030018; PMID: 17274686
- Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, Calderwood SB, Ausubel FM. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A 2001; 98:10892 - 7; http://dx.doi.org/10.1073/pnas.191378698; PMID: 11535834
- Maadani A, Fox KA, Mylonakis E, Garsin DA. Enterococcus faecalis mutations affecting virulence in the Caenorhabditis elegans model host. Infect Immun 2007; 75:2634 - 7; http://dx.doi.org/10.1128/IAI.01372-06; PMID: 17307944
- Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC Jr., Mylonakis E. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc Natl Acad Sci U S A 2008; 105:14585 - 90; http://dx.doi.org/10.1073/pnas.0805048105; PMID: 18794525
- Cruz MR, Graham CE, Gagliano BC, Lorenz MC, Garsin DA. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect Immun 2013; 81:189 - 200; http://dx.doi.org/10.1128/IAI.00914-12; PMID: 23115035
- Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 2004; 54:1212 - 23; http://dx.doi.org/10.1111/j.1365-2958.2004.04349.x; PMID: 15554963
- Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol 2008; 67:47 - 62; http://dx.doi.org/10.1111/j.1365-2958.2007.06013.x; PMID: 18078440
- Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 2001; 67:2982 - 92; http://dx.doi.org/10.1128/AEM.67.7.2982-2992.2001; PMID: 11425711
- Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J 2008; 2:27 - 36; http://dx.doi.org/10.1038/ismej.2007.76; PMID: 18049456
- Wang LH, He Y, Gao Y, Wu JE, Dong YH, He C, Wang SX, Weng LX, Xu JL, Tay L, et al. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol Microbiol 2004; 51:903 - 12; http://dx.doi.org/10.1046/j.1365-2958.2003.03883.x; PMID: 14731288
- Morales DK, Grahl N, Okegbe C, Dietrich LE, Jacobs NJ, Hogan DA. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio 2013; 4:e00526 - 12; http://dx.doi.org/10.1128/mBio.00526-12; PMID: 23362320
- Tampakakis E, Peleg AY, Mylonakis E. Interaction of Candida albicans with an intestinal pathogen, Salmonella enterica serovar Typhimurium. Eukaryot Cell 2009; 8:732 - 7; http://dx.doi.org/10.1128/EC.00016-09; PMID: 19329669
- Peters BM, Jabra-Rizk MA, Scheper MA, Leid JG, Costerton JW, Shirtliff ME. Microbial interactions and differential protein expression in Staphylococcus aureus -Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol 2010; 59:493 - 503; PMID: 20608978
- Pereira-Cenci T, Deng DM, Kraneveld EA, Manders EM, Del Bel Cury AA, Ten Cate JM, Crielaard W. The effect of Streptococcus mutans and Candida glabrata on Candida albicans biofilms formed on different surfaces. Arch Oral Biol 2008; 53:755 - 64; http://dx.doi.org/10.1016/j.archoralbio.2008.02.015; PMID: 18395698
- Jarosz LM, Deng DM, van der Mei HC, Crielaard W, Krom BP. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell 2009; 8:1658 - 64; http://dx.doi.org/10.1128/EC.00070-09; PMID: 19717744
- Bamford CV, d’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun 2009; 77:3696 - 704; http://dx.doi.org/10.1128/IAI.00438-09; PMID: 19528215
- Azoulay E, Timsit JF, Tafflet M, de Lassence A, Darmon M, Zahar JR, Adrie C, Garrouste-Orgeas M, Cohen Y, Mourvillier B, et al, Outcomerea Study Group. Candida colonization of the respiratory tract and subsequent pseudomonas ventilator-associated pneumonia. Chest 2006; 129:110 - 7; http://dx.doi.org/10.1378/chest.129.1.110; PMID: 16424420
- Neely AN, Law EJ, Holder IA. Increased susceptibility to lethal Candida infections in burned mice preinfected with Pseudomonas aeruginosa or pretreated with proteolytic enzymes. Infect Immun 1986; 52:200 - 4; PMID: 2420722
- Harriott MM, Noverr MC. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother 2009; 53:3914 - 22; http://dx.doi.org/10.1128/AAC.00657-09; PMID: 19564370
- Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell 1997; 90:939 - 49; http://dx.doi.org/10.1016/S0092-8674(00)80358-X; PMID: 9298905
- Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2003; 2:1053 - 60; http://dx.doi.org/10.1128/EC.2.5.1053-1060.2003; PMID: 14555488
- Mylonakis E, Engelbert M, Qin X, Sifri CD, Murray BE, Ausubel FM, Gilmore MS, Calderwood SB. The Enterococcus faecalis fsrB gene, a key component of the fsr quorum-sensing system, is associated with virulence in the rabbit endophthalmitis model. Infect Immun 2002; 70:4678 - 81; http://dx.doi.org/10.1128/IAI.70.8.4678-4681.2002; PMID: 12117982
- Qin X, Singh KV, Weinstock GM, Murray BE. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J Bacteriol 2001; 183:3372 - 82; http://dx.doi.org/10.1128/JB.183.11.3372-3382.2001; PMID: 11344145
- Sifri CD, Mylonakis E, Singh KV, Qin X, Garsin DA, Murray BE, Ausubel FM, Calderwood SB. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect Immun 2002; 70:5647 - 50; http://dx.doi.org/10.1128/IAI.70.10.5647-5650.2002; PMID: 12228293
- Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun 2004; 72:4996 - 5003; http://dx.doi.org/10.1128/IAI.72.9.4996-5003.2004; PMID: 15321991
- Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 2011; 9:233 - 43; http://dx.doi.org/10.1038/nrmicro2536; PMID: 21358670
- Mason KL, Erb Downward JR, Falkowski NR, Young VB, Kao JY, Huffnagle GB. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect Immun 2012; 80:150 - 8; http://dx.doi.org/10.1128/IAI.05162-11; PMID: 21986629
- Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, Huffnagle GB. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun 2012; 80:3371 - 80; http://dx.doi.org/10.1128/IAI.00449-12; PMID: 22778094
- Carlson E. Enhancement by Candida albicans of Staphylococcus aureus, Serratia marcescens, and Streptococcus faecalis in the establishment of infection in mice. Infect Immun 1983; 39:193 - 7; PMID: 6401691
- Peters BM, Noverr MC. Candida albicans-Staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect Immun 2013; 81:2178 - 89; http://dx.doi.org/10.1128/IAI.00265-13; PMID: 23545303
- Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 2011; 53:994 - 1002; http://dx.doi.org/10.1093/cid/cir632; PMID: 22002980
- Van Kessel K, Assefi N, Marrazzo J, Eckert L. Common complementary and alternative therapies for yeast vaginitis and bacterial vaginosis: a systematic review. Obstet Gynecol Surv 2003; 58:351 - 8; http://dx.doi.org/10.1097/01.OGX.0000068791.04785.8D; PMID: 12719677
- Liu CM, Hungate BA, Tobian AA, Serwadda D, Ravel J, Lester R, Kigozi G, Aziz M, Galiwango RM, Nalugoda F, et al. Male circumcision significantly reduces prevalence and load of genital anaerobic bacteria. MBio 2013; 4:e00076; http://dx.doi.org/10.1128/mBio.00076-13; PMID: 23592260
- Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife 2013; 2:e00458; http://dx.doi.org/10.7554/eLife.00458; PMID: 23599893
- Project HMC, Human Microbiome Project Consortium. A framework for human microbiome research. Nature 2012; 486:215 - 21; http://dx.doi.org/10.1038/nature11209; PMID: 22699610
- Project HMC, Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207 - 14; http://dx.doi.org/10.1038/nature11234; PMID: 22699609
- Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Kong HH, et al, NIH Intramural Sequencing Center Comparative Sequencing Program. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013; 498:367 - 70; http://dx.doi.org/10.1038/nature12171; PMID: 23698366
- White SJ, Rosenbach A, Lephart P, Nguyen D, Benjamin A, Tzipori S, Whiteway M, Mecsas J, Kumamoto CA. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog 2007; 3:e184; http://dx.doi.org/10.1371/journal.ppat.0030184; PMID: 18069889
- Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 2010; 6:e1000713; http://dx.doi.org/10.1371/journal.ppat.1000713; PMID: 20072605
- Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J 2008; 2:1183 - 93; http://dx.doi.org/10.1038/ismej.2008.76; PMID: 18670396
- Chen Y, Chen Z, Guo R, Chen N, Lu H, Huang S, Wang J, Li L. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis 2011; 70:492 - 8; http://dx.doi.org/10.1016/j.diagmicrobio.2010.04.005; PMID: 20846815
- Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol 2008; 43:831 - 41; http://dx.doi.org/10.1080/00365520801935434; PMID: 18584522
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012; 336:1314 - 7; http://dx.doi.org/10.1126/science.1221789; PMID: 22674328
- Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol 2010; 8:340 - 9; http://dx.doi.org/10.1038/nrmicro2313; PMID: 20348933
