Abstract
As a leading cause of congenital infection and a major threat to immunocompromised individuals, human cytomegalovirus (HCMV) is a major global public health concern. Effective HCMV vaccines would need to induce potent and balanced humoral and cellular immune responses. In this pilot study, immunogenicity studies were conducted in mice to examine HCMV antigen-specific antibody and T cell responses when a heterologous prime-boost immunization strategy was tested. DNA vaccines expressing either targets of protective antibody responses (gB and gM/gN) or well characterized T cell immunogens (pp65, pp150, and IE1) were used as the priming immunization while the live attenuated HCMV vaccine Towne strain was used as the boost, which may act like an inactivated vaccine due to the inability of HCMV to replicate in a mouse host. Our data indicate that while DNA vaccines were effective in priming HCMV-specific antibody responses, the final titers of gB- or gM-specific antibodies were not much different from those elicited by using multiple immunizations of HCMV alone. In contrast, DNA priming significantly enhanced T cell responses against gB, pp65, and IE1 as measured by IFN-γ. However, HCMV alone was not effective in eliciting strong T cell immune responses when used in a mouse host. Our data indicate that the complexity of antigen composition from a large virus, such as HCMV, may affect the profile of immune responses when viral vaccines are used as a boost.
Introduction
Human cytomegalovirus (HCMV) is a β-herpesvirus characterized by its restricted host range. It is the largest known human herpesvirus, with a genome of about 230 kb. The virus has double-stranded linear DNA mixed with a matrix protein (the tegument) surrounded by a lipid bilayer containing viral glycoproteins.Citation1 A primary infection is likely to be followed by lifelong persistence of the virus in a latent phase and clinical symptoms are seen when the immune system is not able to control the virus. Therefore, the interest in HCMV as a major pathogen to humans has remained high given its threat to immunocompromised individuals, such as recipients of hematopoietic cell transplant (HCT), solid-organ transplants, as well as HIV-infected patients. Moreover, congenital HCMV infection is one of the leading causes of severe morbidity and mortality in newborns.Citation2
In the year 2000, the Institute of Medicine identified HCMV vaccine development as a high priority due to the lifelong and severe morbidity of newborns who develop symptomatic congenital disease in utero.Citation3 Over the years, many attempts were made to develop an efficacious protective CMV vaccine. Different strategies, either used alone or in combination, have been clinically evaluated, including live attenuated virus, subunit vaccines, and viral vectors encoding different HCMV immunogens. Administration of Towne strain HCMV provided partial protection against severe disease in transplant recipients but was unsuccessful in protecting against infection in children who were exposed to pregnant mothers who shed the virus.Citation4 Although preliminary reports demonstrated safety of a Towne/Toledo chimera vaccine, efficacy studies have never been attempted.Citation5
The envelope glycoprotein B (gB) of HCMV has been used as the sole viral antigen in subunit and vector-based vaccines, mainly to generate neutralizing antibodies against the virus. Although gB-based subunit vaccines were well tolerated and induced a high level of antibodies, there were no significant differences between recombinant CMV gB vaccine and placebo recipients in a randomized, double-blind, Phase II clinical trial of seronegative woman of childbearing age.Citation6 These results raised doubt whether using one of the envelope glycoproteins alone as a vaccine would induce broad enough neutralizing antibody responses to prevent intrauterine transmission in all instances. Using non-human primates (NHP), a vaccine composed of HCMV subunits gB, pp65, and IE1 was found to be partially effective at preventing shedding using a virulent Rhesus CMV challenge model.Citation7 The discovery of a second entry portal, applicable to both human and Rhesus CMV, has led to a revolution in vaccine design in which subunits from a complex of proteins, the UL128 complex, has been shown to be necessary for entry into epithelial/endothelial cells. A Rhesus CMV vaccine composed of all five subunits of the UL128 complex pentamer provided partial protection against a virulent endotheliotropic strain of Rhesus CMV.Citation8 These studies point out the likelihood of needing vaccine solutions for both the fibroblast and epithelial/endothelial portals of CMV entry to create an effective prophylactic vaccine.
T cell immune responses have been proposed as important for the efficacy of HCMV vaccines, with the main focus being on two T cell antigens, pp65 and IE1.Citation9,Citation10 Canarypox vector expressing pp65 induced a strong cell-mediated response in addition to antibody responses and the frequency of CD8 T cells was comparable with that of unvaccinated, naturally infected CMV-positive donors.Citation11 The combination of gB/pp65/IE1 as antigens used in a replication-deficient alphavirus vector was evaluated in a Phase 1 clinical trial and results showed that the magnitude of the T cell response (based on IFN-gamma production) was greatest for pp65 followed by gB and IE1 antigen.Citation12 Moreover, a significant proportion of CMV-specific T cell responses showed polyfunctional phenotypes thought to be crucial for protective immune responses.Citation13
An important new platform used to develop a vaccine against HCMV is DNA immunization.Citation14 DNA vaccines, based on gB, pp65 and IE1 antigens (trivalent vaccine) or gB/pp65 (bivalent DNA vaccine), have been evaluated in human clinical studies; however, the immunogenicity of DNA vaccines was low when tested alone in humans. In recent years, the idea of maximizing the immune response through sequential administration of vaccines that use different antigen delivery systems (“heterologous prime-boost”) has attracted significant attention.Citation15-Citation17 It has been shown that such an approach is effective in enhancing immunity and enriching T cells with high avidity to particular immunogens and would improve the level of protection against infection.Citation18,Citation19 One of the first approaches to utilize this heterologous prime-boost regimen was made in the early 1990s in non-human primate models by immunization with recombinant vaccinia virus encoding the gp160 antigen of SIVmne, which was followed by boost with recombinant gp160 protein expressed from baculovirus-infected cells.Citation20 Following this immunization regimen, animals were protected from challenge with the SIVmne virus. The efficacy of this heterologous prime-boost approach has also been demonstrated in studies of HIV-1, malaria, and tuberculosis.Citation21-Citation23 This approach was also shown to improve the effectiveness of existing vaccines, e.g., against infection with rabies or hepatitis B virus (HBV).Citation24,Citation25
In the current study, we tested whether DNA priming is effective in eliciting HCMV-specific antibody and T cell immune responses in a mouse model when HCMV viral stock is used as the boost. Groups of mice immunized with either DNA alone or HCMV viral stock alone were used as the controls. Results from this study will help identify the optimal prime-boost combination to maximize immune responses against HCMV.
Results
Construction and evaluation of DNA vaccines expressing HCMV antigens
In the current study, a group of DNA vaccines were constructed to express individual candidate HCMV antigens. The primers used in cloning these HCMV genes are described in . HCMV strain AD169 was used as the source to clone the above antigen genes for the construction of DNA vaccines. The expression of HCMV gene DNA vaccines was tested in a transient transfection system in 293T cells and verified by western blot analysis.
Table 1. Primers to amplify the gB, pp65, pp150 and IE1.4 genes from HCMV genome to construct individual DNA vaccines
The first DNA vaccine expresses HCMV protein UL55 (gB), which is a well characterized target of protective antibodies against HCMV. The UL55 gene encodes the full length (907 amino acids) gB protein, which is further glycosylated and is expected to have a molecular weight of 170 kD ().Citation26 The gB protein can be further cleaved intracellularly, into two subunits, gp116 and gp55. Like their counterparts in other enveloped viruses, such as HIV-1 and SARS-CoV, the transmembrane subunit gp55 anchors the gB protein on the surface of HCMV viral particles. We have previously shown that truncating the transmembrane subunits by removing transmembrane sequences and intracellular regions could lead to increased secretion and immunogenicity of HIV-1 and SARS-CoV envelope proteins when they were expressed by DNA vaccine plasmids.Citation27,Citation28 In the current study, a similarly truncated gB DNA vaccine for HCMV (gB-s) was also constructed, which removed the C-terminal 277 aa starting just upstream of the transmembrane region (). Western blot results () showed that a gB-s design led to significant amounts of secreted, truncated gB protein, which is expected to have a molecular weight of about 140 kD, in addition to those that remain in the cell lysate. Cleavage of this truncated gB produced a 30 kD protein (remaining portion of the original gp55 transmembrane subunit) and the gp116 subunit. Interestingly, secreted gB-s showed less cleavage than gB-s in cell lysate. Furthermore, the full length gB, only detectable in cell lysate, mainly produces the gp55 and a number of lower molecular weight bands but not the distinctive gp116 unit.
Figure 1. Design of HCMV glycoprotein B DNA vaccine constructs with different forms of inserts. Schematic representation of the full length gB protein (amino acid 1–907) is shown at the top and DNA vaccine expressing truncated just prior the transmembrane region of gB (amino acid 1–692) is shown in the lower part of the figure (A). Expression of gB-full length and shorted gB in HEK 293 T cells (Western Blot analysis) (B). To detect of gB antigen rabbit sera collected at 1 week after forth immunization with gB-full length and gB-s were used. Cell lysates (L) and supernatants (S) from 293T cells transfected with two different gB constructs and the empty vectors (negative control) are labeled above each western blots. Molecular weight was indicated on the right. Comparison of neutralizing activity of rabbit antisera against AD169 strain of HCMV (C). Rabbits were immunized with plasmid DNA administered to individual animal at weeks 0, 2, 4, 6. Sera tested were forth bleeding from rabbit immunized with DNA vaccines: pJW4303 (negative control), pJW4303/gB and pJW4303/gB-s. 2-fold serial dilutions of antisera were incubated with 100 pfu of AD169 virus before the mixture was used to infect human primary fibroblasts. Neutralization titers shown were the highest rabbit sera dilutions at which 50% reduction of HCMV infection was achieved.
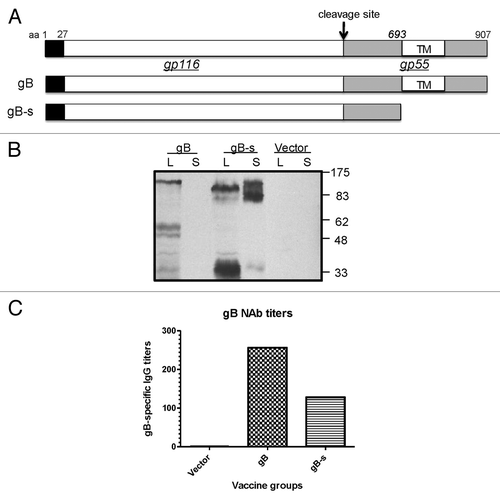
The immunogenicity of these two versions of gB DNA vaccines were tested in rabbits. The full length gB DNA vaccine elicited higher neutralizing antibodies than the gB-s DNA vaccine (). Therefore, in the current study, the full length gB DNA vaccine was used as part of the polyvalent DNA vaccine formulation.
Another HCMV envelope glycoprotein pair, UL100 (gM) and UL73 (gN), was also used in the current study. The construction and expression of gM and gN DNA vaccines was previously described. Their immunogenicity was tested in animal studies and it was shown that they elicited protective antibodies against HCMV.Citation29
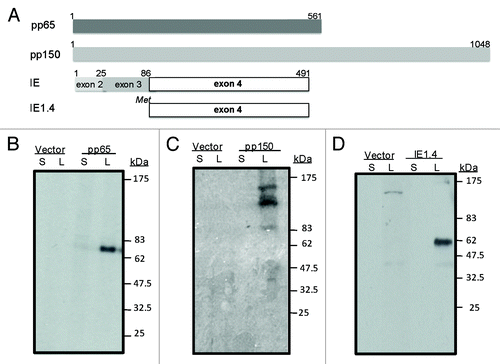
Two HCMV tegument proteins, UL83 (pp65), and UL32 (pp150), encoding the 561 aa and 1048 aa phosphorylated proteins, respectively, are the main targets of HCMV T cell immune responses.Citation28 The HCMV UL123 or the immediate early 1 (IE1) protein is another main target for T cell immune responses.Citation30,Citation31 IE1 is composed of four exons, among which, exon 4 (IE1.4) is the most immunogenic.
The designs of pp65, pp150, and IE1/exon 4 DNA vaccines are shown in and the expression of these DNA vaccines was confirmed in transient transfected 293T cells and verified by western blot analysis (). Expression of pp65, pp150, and IE1.4 proteins was clearly detected but only in cell lysate. As a negative control to HCMV DNA vaccines, the empty DNA vector, pJW4303, was included in all analyses. The expression of the pp65 or IE1.4 proteins showed a single band of expected molecular weight for both antigens. Expression of the pp150 protein showed a 150 kD band but also several additional bands of lower molecular weight, suggesting significant intracellular degradation of the pp150 protein.
Figure 2. Schematic design of plasmid DNA constructs (A) and expression of pp65, pp150 and IE1.4 proteins in HEK 293 T cells (Western Blot analysis) (B-C) and (D) respectively). At the top we presented pp65 plasmid DNA construct (amino acid 1–561) representing full length protein; plasmid DNA construct encoding entire pp150 protein (amino acid 1–1048) was shown in the middle section and in the lower part plasmid encoding exon 4 of IE1 protein (amino acid 86–491) was presented (A). Antigen expression by pp65 and pp150 DNA vaccines was detected by pooled murine antiserum from group immunized with DNA-pp65 (B) or group immunized with DNA-pp150 vaccine (C), respectively. Expression of IE1.4 DNA vaccine construct was detected by murine monoclonal antibody, mAb p63–27 (D). Presence of pp65, pp150 and IE1.4 antigens was analyzed in supernatants (S) and lysates (L) from 293T cells. Supernatant and lysate from 293T cells transfected with vector alone was used as negative control. Molecular weight was indicated on the right.
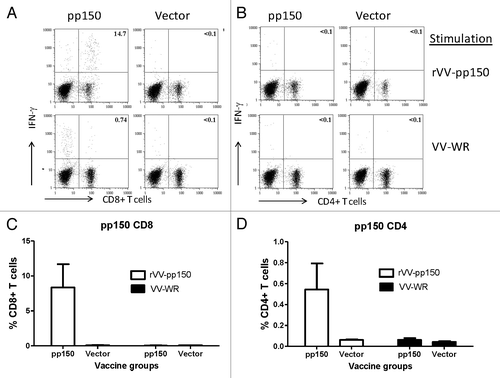
Immunogenicity of these T cell immunogens was tested individually in mice. ELISpot with splenocytes from immunized mice showed significant levels of pp65- or pp150-specific T cell responses when stimulated with vaccinia viruses expressing respective HCMV antigens while cells stimulated with regular vaccinia viruses or mice immunized with empty vector did not show pp65- or pp150-specific T cell responses (). Intracellular cytokine staining (ICS) was also conducted to measure CD4+ and CD8+ T cell responses elicited by these two DNA vaccines. After stimulation with pp65-expressing VV, mice immunized with pp65 DNA vaccine showed high level pp65-specific CD8+ T cell responses (> 4%) and strong CD4+ T cell responses (> 1%) (). In mice immunized with pp150 DNA vaccine, the CD8+ T cell responses were much stronger (~8%) than the CD4+ T cell responses (just over 0.5%) (). Stimulation with VV-WR only did not show a significant antigen-specific T cell response ( and ).
Figure 3. Production of IFN-gamma tested by ELISpot in splenocytes received from mice immunized with DNA-pp65 vaccine alone (A) or DNA-pp150 vaccine alone (B). Mice were immunized with 3x DNA vaccine encoding pp65 or pp150 antigens. Splenocytes from group of mice immunized with DNA-pp65, DNA-pp150 or vector alone were stimulated with rVV-pp65 (A) or rVV-pp150 (B). As control all immunization groups were stimulated with VV-WR.
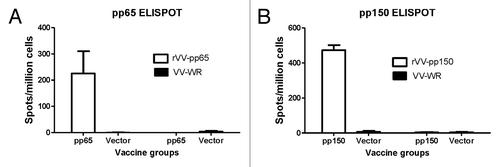
Figure 4. T cell response in mice immunized with DNA-pp65 vaccine alone tested by ICS assay. Representative dot plots show the percentage of IFN-g positive CD8+T cells (A) and IFN- g positive CD4+T positive cells (C). Lymphocytes gated on CD3+ T cells were further gated on CD8+ or CD4+ T lymphocytes. Splenocytes were stimulated with rVV-pp65 (top panel) and with VV-WR (bottom panel) (A) and (C). The levels of specific CD8+T cells are shown as percentage of IFN-gamma positive CD8+T cells (B) or IFN-gamma positive CD4+ T cells (D) in response to rVV-pp65 or VV-WR in two immunization groups (mice immunized with DNA pp65 vaccine or vector alone vaccine).
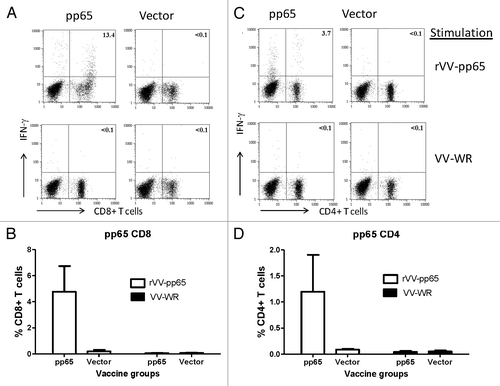
Figure 5. Induction of pp150 CD8+T lymphocytes and CD4+T cell lymphocytes following immunization with DNA pp150 vaccine alone. Representative analysis of intracellular cytokine staining assay on splenocytes stimulated with rVV-pp150 (top panel) and VV-WR (bottom panel) was shown for CD8+T cell population (A) and CD4+ T cell population (C). Numbers on the CD8+ and CD4+T–lymphocyte plots denote the percentage of the population expressing IFN-gamma (A and C). Lymphocytes gated on CD3+ T cells were further gated on CD8+ or CD4+ T lymphocytes. The levels of specific CD8+T cells in two immunization groups are demonstrated as percentage of IFN-gamma positive CD8+T cells (B) or IFN-gamma positive CD4+ T cells (D) in response to rVV-pp150 or VV-WR.
Designs of prime-boost studies
The above HCMV DNA vaccines were further grouped as two priming immunization formulations and tested in a pilot prime-boost study to compare the relative immunogenicity between the homologous and the heterologous prime-boost approaches (). One priming formulation included gB/gM/gN DNA vaccines, which focused on the induction of antibody responses against HCMV antigens; the other formulation included pp65/pp150/IE1.4 DNA vaccines as the prime, which focused on the induction of T cell immune responses. Within each formulation, equal amounts of three DNA plasmids were used and the DNA immunizations were delivered via gene gun at Weeks 0 and 2 as the priming immunization or Weeks 0, 2, and 4 as the full series of immunization.
Figure 6. Immunization groups and DNA vaccines components. At week 0 and 4 mice were primed with the following vaccines: CMV alone, vector alone, DNA-1 alone, DNA-1+ CMV, DNA-2 alone or DNA-2+ CMV. At week 4 animals were boosted with either CMV or DNA-1 or DNA-2 immunizations.
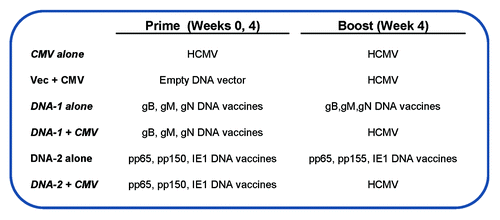
A live attenuated HCMV vaccine made using the Towne strain was tested in the current study as the boosting vaccination and was delivered intraperitoneally (i.p.). Because HCMV does not replicate in mice, this viral stock functioned similarly as an inactivated vaccine and is expected to boost HCMV-specific immune responses based on the HCMV protein antigens contained in this HCMV viral stock. For two key testing groups (), HCMV vaccine was used to boost mice primed with either the gB/gM/gN DNA formulation or the pp65/pp150/IE1.4 DNA formulation (“heterologous prime-boost”). Three other groups received three times either DNA alone or HCMV alone immunizations (“homologous prime-boost”). The last group is a control group that received the empty DNA vector as the prime and HCMV as the boost.
Antibody responses elicited by heterologous prime-boost immunization
Mouse serum antibody responses were measured by ELISA in groups of mice immunized with a gB/gM/gN DNA vaccine combination with or without boost with HCMV viral stock (). The selection of gB and gM proteins as antigens was based on previous studies showing them as the main targets for neutralizing antibodies.Citation29,Citation32,Citation33 Due to the small amount of mouse sera, we did not perform a microneutralization assay and instead, used the levels of binding antibodies to reflect the effectiveness of prime-boost immunizations. For gB antibody responses, recombinant gB () produced in the lysate of 293T cells was used as the coating antigen while, for measurement of gM antibody responses, a synthetic peptide representing the known key gM epitope was used as the coating antigen.
Figure 7. Detection of antibodies specific to gB (A) and gM (B) antigens by ELISA. gB-specific and gM- specific IgG titers were measured in mice sera collected after: 3 immunizations with DNA-1 alone, 2 immunizations with DNA-1 plus CMV, 3 immunizations with CMV alone or 2 immunizations with vector alone plus CMV. Data are shown as geometric mean titers within the group. P values indicate statistically significant differences measured by T test.
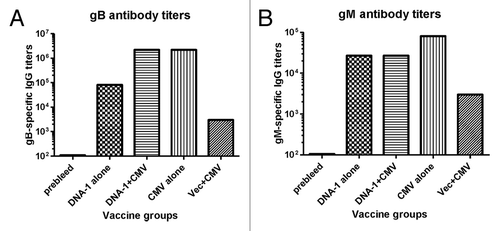
Three-time DNA alone immunizations elicited high levels of gB-specific antibody responses (), and twice DNA priming immunizations followed by one-time HCMV boost was able to further increase the antibody responses by another 1.5 logs. However, three-time immunization with HCMV was also able to reach the same high-level antibody responses elicited by the DNA prime-HCMV boost but did not further increase levels, indicating the effectiveness of DNA as a prime but not as a boost. In contrast, one-time HCMV immunization following twice empty DNA prime was not able to elicit high-level gB antibody responses, suggesting that the HCMV DNA priming effect was antigen-specific and was not based on a non-specific DNA adjuvant effect.
A similar pattern of antibody responses was observed against the gM antigen but the overall level of antibody responses against gM were low relative to antibodies against the gB antigen. Three times DNA immunization alone was not able to elicit significant gM antibody responses, nor was one-time HCMV immunization after empty DNA vector priming (). Only DNA vaccine prime followed with HCMV boost or repeated immunizations with HCMV was able to elicit high-level anti-gM antibody responses. Our previous report showed that the immunogenicity of the gM DNA vaccine was relatively low based on antibody titers.Citation29 Furthermore, in the current study, only antibodies against a key epitope of gM were examined, which may be lower than the antibodies against the entire gM protein. However, the expression level of gM by transient expression system is too low to provide a sufficient amount of gM proteins for ELISA studies. We did not measure antibody responses against gN antigen alone as both literature and our previous report showed that the main role of gN is to be part of the gM/gN immunogen and it is difficult to detect antibodies against gN alone.Citation29,Citation33
T cell responses elicited by heterologous prime-boost immunization
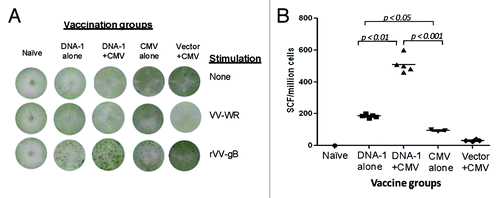
In this pilot study, we used IFN-gamma ELISPOT as the biomarker to measure HCMV-specific T cell immune responses after splenocytes from immunized mice were stimulated in cultured medium with vaccinia virus expressing respective HCMV antigens. Compatible to anti-gB antibody responses, significant levels of gB-specific T cell immune responses were detected in the group of mice that received three-time immunizations with the gB/gM/gN DNA vaccine formulation (). Furthermore, a 2.5-fold increase in T cell immune responses was observed in the group of mice that received a boost of HCMV after two-time priming immunizations with DNA vaccines (). However, different from gB antibody responses, three-time immunizations with HCMV did not elicit high levels of gB-specific T cell immune responses, and the responses were only slightly higher than those observed in mice that received one-time HCMV immunization after empty DNA vector prime. Naïve mice did not show positive IFN-gamma ELISPOT responses even after stimulation by gB-vaccinia viruses and stimulation with the VV-WR did not show antigen-specific T cell immune responses.
Figure 8. T cell response specific to gB antigen. Detection of gB-SCs producing IFN-gamma measured by ELISpot assay (A) and enumeration of gB-SCs (B). An example of the spots generated in response to VV-gB is represented for four groups of mice immunized with: 3x DNA-1 alone (gB/gM/gN), 2x DNA-1 plus CMV, 3x CMV-alone and 2x vector alone plus 1x CMV. As negative control splenocytes from naïve mouse were used (A). The mean numbers of antigen-specific spot forming cells after background subtraction of control wells with no antigen were plotted (B). Experiments were conducted in triplicate. Data are shown as geometric mean titers within the group. P values indicate statistically significant differences measured by T test.
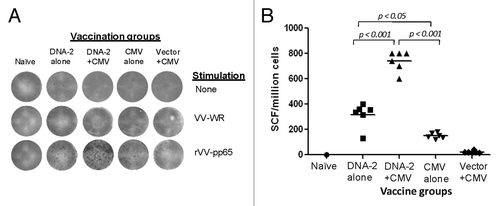
Additional IFN-gamma ELISPOT analyses were conducted in mice that received the pp65/pp150/IE1.4 DNA vaccine combination. T cell responses to pp65 were very similar to gB-specific T cell responses (). DNA vaccine alone was able to elicit high-level pp65-specific IFN-gamma responses while the DNA prime-HCMV boost elicited the highest levels of T cell responses. Similar to gB T cell responses, HCMV vaccine alone was not able to elicit high-level T cell responses, and responses were only slightly higher than a one-time HCMV immunization after empty DNA vector prime.
Figure 9. IFN-gamma producing T cell response specific to pp65 antigen. Detection of pp65-SCs producing IFN-gamma measured by ELISpot assay (A) and enumeration of pp65-SCs (B). Representative example of spots generated to VV-pp65 in four groups of mice immunized with 3x DNA-2 alone (pp65/pp150/IE1.4), 2x DNA-1 plus CMV, 3x CMV-alone and 2x vector alone plus 1x CMV. Splenocytes from naïve mouse were used as negative control. The mean numbers of antigen-specific spot forming cells after background subtraction of control wells with no antigen were plotted (B). Experiments were conducted in triplicate. The student T test was used to compare frequencies between groups and p values are depicted in the panel.
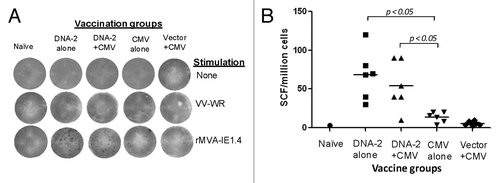
The overall IFN-gamma ELISPOT response to another T cell antigen, IE1.4, was lower than observed for gB- or pp65-specific IFN-gamma ELISPOT responses (). Here, DNA vaccine alone was as immunogenic as the DNA prime-HCMV boost approach in eliciting IE1.4-specific T cell responses. Again, three-time immunization with HCMV alone was not very immunogenic, as it elicited responses that were almost similar to those elicited by one-time immunization with HCMV after empty DNA vector prime.
Figure 10. Cell mediated response against IE1.4 antigen measured by production of IFN-gamma in mouse splenocytes. Detection of IE1.4-SCs producing IFN-gamma measured by ELISpot assay (A) and enumeration of IE1.4-SCs (B). An example of the spots generated in response to VV-IE1.4 is represented for four groups of mice immunized with: 3x DNA-1 alone (pp65/pp150/IE1.4), 2x DNA-1 plus CMV, 3x CMV-alone and 2x vector alone plus 1x CMV. As negative control splenocytes from naïve mouse were used (A). The mean numbers of antigen-specific spot forming cells after background subtraction of control wells with no antigen were plotted (B). Experiments were conducted in triplicate. The student T test was used to compare frequencies between groups and p values are depicted in the panel.
Unexpectedly, no significant IFN-gamma ELISPOT response was detected in mice that received either the pp65/pp150/IE1.4 DNA vaccine combination (with or without HCMV boost), or HCMV alone vaccine when stimulated with rVV-pp150 (data not shown).
In contrast to clearly positive IFN-gamma responses detected after stimulation with vaccinia viruses expressing HCMV antigens, stimulating splenocytes from immunized mice with VV-WR did not lead to positive detection of IFN-gamma responses for gB, pp65, or IE1.4 antigens (–). provides a summary of HCMV-specific IFN-gamma ELISPOT T cell responses detected in the current study.
Table 2. Summary of HCMV antigen –specific T cell response in immunized mice
Discussion
An effective CMV vaccine would be able to protect human populations in special circumstances. While the previously licensed live attenuated HCMV vaccine based on Towne’s strain could elicit neutralizing antibodies and T cell responses, its wide use was limited due to inadequate antigen-specific interferon gamma responses by CD4 and CD8 T cells following vaccination.Citation34-Citation36 The fact that a HCMV vaccine induced an immune response similar to natural infection suggests that the Towne vaccine may be over-attenuated as a result of extensive passage in cultured cells. To improve efficacy of a live HCMV vaccine, to make it more immunogenic that the Towne vaccine, genetic recombinants of Towne/Toledo strains were constructed, but they failed to boost humoral or cellular immune responses in seropositive volunteers.Citation5 It is now understood that HCMV strains that are extensively cultured on fibroblasts lose the endocytic tropism by deletion of members of the UL/b’ complex.Citation37 These strains no longer have endocytic tropism and are not actively shed or have the virulence of natural isolates which possess the UL/b’ region. Reconstitution of bacterial artificial chromosome forms of Rhesus or human CMV which have deletions in the UL/b’ region were shown to restore virulence and the capacity to infect through the endocytic pathway which further demonstrates the importance of this region for virulence.Citation38 Landmark studies caused a reinterpretation of classical thought processes regarding immunodominance of the fibroblast pathway by demonstrating that if a suitable alternative test cell line is used, neutralizing antibodies against the endocytic pathway predominate in human serum from most CMV-seropositives.Citation39
To improve the efficacy of any candidate HCMV vaccine against the largest human viruses ever known is a big challenge. More than one immunogen may be needed to provide complete protection against infection, especially against the wide range of primary viral isolates that have been identified in recent years.
In addition to traditional live attenuated or inactivated vaccine approaches, which are developed based on a particular strain of HCMV, various novel approaches have been explored to include only a few common antigens of HCMV. HCMV vaccines based on recombinant gB protein have been tested in humans and were shown immunogenic in eliciting gB-specific antibody responses. At the same time, recombinant protein-based vaccines are not the ideal candidates for the induction of T cell immune responses.
DNA vaccination technology, discovered in the early 1990s, is known for its potential to elicit T cell immune responses while delivering multiple antigens at the same time.Citation40 DNA vaccines expressing selected HCMV antigens have been tested in humans,Citation41 however, their immunogenicity in humans was poor, similar to reports of other human DNA vaccine studies.Citation42 Since 2008, several groups have demonstrated that DNA vaccines are effective in priming a host’s immune system, including humansCitation21,Citation43,Citation44 if a follow up boost, in the form of protein or viral vector, is delivered. This type of “heterologous prime-boost” vaccination strategy has been shown even with non-DNA vaccination approaches.Citation17
In the current report, we conducted a pilot study to use the DNA prime-viral stock boost approach to test its relative effectiveness in delivering two multi-gene HCMV DNA formulations as the priming immunization while using the homologous viral stock prime-boost approach as the control. The HCMV viral stock selected in the current study is the Towne strain of HCMV, which is the same as the licensed live attenuated HCMV vaccine. While HCMV viral stock is not able to replicate in a mouse host and, thus, its immunogenicity is reduced in contrast to what could be achieved in a permissive host, the use of this viral stock in the current study can still provide a full spectrum of HCMV antigens, similar to “inactivated” vaccines. Previous reports, including those from our group, demonstrated that DNA vaccines were effective in priming host immune responses, which can be further boosted by inactivated rabies or influenza vaccines.Citation24,Citation45,Citation46 We used one HCMV DNA formulation with known targets of protective antibodies (gB/gM/gN) to prime antibody responses and the other HCMV DNA formulation, with well-studied T cell immunogens (pp65/pp150/IE1.4), to focus on T cell responses. The same HCMV viral stock was used as the boost for both types of DNA priming and revealed interesting differences between antibody and T cell responses against key HCMV antigens.
Our results confirmed previous knowledge learned from HIV and influenza vaccines studies that DNA immunization is effective in priming antibody responses. Because twice DNA prime and one-time HCMV viral stock boost was able to elicit the same high-level of antibodies as three-time HCMV viral stock immunizations, while one-time HCMV viral stock was not sufficient to do so; DNA priming is thus equally effective as priming by HCMV viral stock. However, in our previous HIV or influenza vaccine studies, the heterologous prime-boost was more effective than the homologous prime-boost. The difference may be related to the high number of antigens included in the HCMV viral stock, preventing the boost from being “focused.” There is, therefore, a competition against various HCMV antigens that was not included in the priming formulation thus, reducing the power of boost.
On the other hand, T cell immune responses were significantly elevated in the heterologous prime-boost groups (against gB and pp65). This may imply that DNA immunization is more effective than inactivated vaccines in establishing T cell memory responses, as it was hypothesized when DNA vaccination was originally discovered, and supported by the results in the current study that DNA vaccine alone was more immunogenic in inducing T cell responses than HCMV alone. It is still quite impressive to see that the HCMV viral stock, a poor T cell inducer due to its inability to replicate in the hosts, could boost T cell immune responses elicited by DNA priming immunization. Understanding the exact mechanism for this boosting effect will expand potential applications for inactivated vaccines no matter whether priming is delivered by DNA vaccines or not.
In the current study, relative levels of antibody or T cell immune responses varied. This is not completely unexpected as the immunogenicity of individual HCMV antigens may be different even if they are all delivered by DNA immunization. In addition, one weakness of the current study is that we could not effectively determine the relative amounts of individual antigens in the HCMV viral stock, which could also affect the boosting effects. However, our results are quite reassuring as antibody responses and T cell immune responses showed unique patterns (no matter what antigen was studied) when either homologous or heterologous prime-boost approaches were tested.
As a pilot study, we attempted to capture the key signals in a heterologous prime-boost vaccination strategy when HCMV DNA vaccines are used as the prime; we did not, however, conduct extensive analysis on additional immunogenicity parameters. No functional antibodies were tested in the current study due to the limited amounts of immune sera obtained from a mouse study. Because human CMV antigens were tested, it was not possible to conduct a protection study in the mouse model. Only IFN-gamma ELISPOT was used as the representative T cell immune response parameter; however, we are aware that it is possible that other T cell cytokines may show a different profile.
Similar heterologous prime-boost CMV vaccines studies were reported with different study designs. Morello et al. tested a mouse CMV (MCMV) vaccine in the mouse model using multiple DNA plasmid prime and inactivated MCMV boosting strategies.Citation47 As a result, protective immunity against mucosal and systemic virus challenge was observed. Similar results were also shown in another study where priming immunizations with trivalent DNA (IE1, M84-pp65 and gB) and further boosting with inactivated MCMV generated higher levels of pp65-specific CD8 T cells than T cells directed against the IE1 protein.Citation48 Moreover, in this heterologous prime-boost regimen, the authors observed an outstanding humoral response and eventually, high levels of protection against systemic and mucosal challenge with MCMV. One interesting result of our study was that the recall T cell response to IE1.4 in mice immunized with DNA vaccine alone was comparable to that generated after boost with Towne virus. It was previously demonstrated that the Towne vaccine mostly induces sustained IE-dominant T cell responses and low IFN-gamma responses to pp65 stimulationCitation36 which contradicts what is found in natural HCMV infections. Therefore, it was surprising that our DNA prime-Towne virus boost formulation simultaneously enhanced T cell response specific to pp65 antigen and induced detectable IE-specific T cell responses. Jacobson et al. showed that individuals primed with a DNA vaccine and boosted with Towne strain also had lower T cell responses against IE1 than those primed-boosted with the Towne strain alone.Citation49 We also presume that weaker cellular responses against IE1 may be explained by the following facts: (1) pp65 protein, as a highly immunodominant antigen, could suppress the T cell response to IE1.4 antigen; (2) IE1 is a less abundant virion component (< 0.1% of total virion proteins) as opposed to pp65 (15.4%); (3) in vaccine development, application of replication deficient virus (injections of mice with HCMV) excludes the possibility of production of proteins that are usually highly immunogenic in the host after natural infection; (4) we decided to utilize only part of the IE1 protein, which, even though it contains highly immunogenic epitopes, does not include all other epitopes that can also stimulate the immune system.Citation9,Citation50 The protective efficacy of a pp65-specific response is still unknown, however, it was previously shown that strong CD8 T cell responses, specific to IE1, correlated with protection from CMV disease in solid organ transplant recipients.Citation31 Therefore, it is critical to include this antigen in future CMV vaccine development.
In summary, our results confirmed the importance of priming the immune system with DNA vaccines to induce strong cellular and humoral responses against HCMV, in addition to the value of boosting with a large virus even though the focus of the boost may vary depending on the antigen content in HCMV. Furthermore, considering the fact that we were able to improve the efficacy of the DNA vaccine without application of adjuvants or immunomodulating materials speaks to the importance of the heterologous prime-boost strategy in vaccine development. Application of the whole virus guarantees that such an approach distributes protective antigens and natural “built-in” adjuvants. Despite the fact that DNA vaccines are less immunogenic in humans, here we prove their potential as effective priming immunogens.Citation17
Materials and Methods
Cells and viruses
The AD169 strain of HCMV was purchased from American Type Culture Collection (#VR-538) whereas Towne strain was kindly provided by Dr. E. Mocarski (Emory Vaccine Center). Viruses were propagated in primary human foreskin fibroblast cells (FSK) purchased from American Type Culture Collection (#CCD-111sk) following the protocol provided by Dr. E. Mocarski. The AD169 strain was used for DNA construct preparations and neutralization assay and Towne strain (live attenuated) was applied for immunizations.
Recombinant vaccinia virus stocks expressing HCMV antigens were provided by Dr. L. Gibson (University of Massachusetts Medical School) by MTA from one of the authors (DJD) as follows: (rVV-gB,Citation51 rVV-pp65,Citation52 rVV-pp150,Citation53 rMVA-IE1.4.Citation54 Western-restricted (WR) strain of vaccinia virus (VV-WR), used as a positive control in ELISpot and ICS assays was kindly provided by Dr. Lisa Selin, University of Massachusetts Medical School.
HCMV DNA vaccine constructs
Genes coding for HCMV genes (gB, gM, gN, pp65, pp150, and IE1.4) were PCR amplified with pfu polymerase (Stratagene #600136) and cloned into previously described pJW4303 vector.Citation55 The sequences of all primers applied in this study are shown in . The genes encoding gB-full length, pp65, and pp150 antigens were amplified directly with indicated primers. The gene encoding the gB-s fragment was first amplified with pair of primers specific for full length gB: CMV gB-1/CMV gB-2 and then re-amplified with CMV gB-1/CMV gB-3 primers. The gene encoding IE1.4 was amplified with semi-nested PCR using CMVIE1–1 and CMVIE1–2 for the first round of amplification. For re-amplification, primers CMVIE1–3 and CMVIE1–2 were used. DNA vaccine constructs expressing either gM or gN antigens were previously described.Citation29 All prepared inserts were subsequently digested with restriction enzymes HindI III and BamH I or NheI and BamHI and then ligated into to the corresponding sites in the DNA vaccine vector. Correct DNA vaccine clones were confirmed by restriction enzyme analysis and large DNA preps were purified using the Mega plasmid purification kit (Qiagen #12181).
Western Blot analysis
In vitro expression of HCMV antigens by individual DNA vaccines included in the current study was confirmed by transient expression in 293T cells and verified by western blot analysis as previously described.Citation29,Citation56 For detection of gB antigen rabbit serum, samples collected at 1 week after the fourth DNA immunization (36 μg/36 shots/immunization) with gB-full length and gB-s were used (). Mouse sera collected after the fourth DNA immunization with pp65 (6 μg/6 shots/immunization) and pp150 (6 μg/6 shots/immunization) were used to detect pp65 () and pp150 () antigens. Monoclonal antibody p63-27 was kindly provided by Dr. W. Britt (University of Alabama) and was applied for detection of IE1.4 antigen ().
Animal immunization
Female BALB/c mice, 6–8 weeks of age, were purchased from Taconic Farms and housed in the facility of Department of Animal Medicine at the University of Massachusetts Medical School (UMMS). Animal care and immunization studies were conducted in accordance with UMMS IACUC approved protocols.
Each animal group included five mice. To deliver the DNA vaccines, animals were immunized with a Helios gene gun (Bio-Rad Laboratories #165–2431) at the shaved abdominal skin as previously reported.Citation57 Each mouse received two or three bi-weekly immunizations with 6 μg of plasmid DNA (2 µg/each DNA vaccine in both gB/gM/gN and pp65/pp150/IE1.4 formulations) per immunization. For those mice that received live attenuated HCMV as the vaccine, they were immunized i.p. with 106 pfu of HCMV Towne strain in 0.2 ml of medium. The control injection with empty DNA vector (6 μg) was delivered by a gene gun. Blood samples were collected peri-orbitally before the first immunization and 2 weeks after each immunization. Mouse splenocytes were collected 2 weeks after the third immunization.
Enzyme-linked immunosorbent assay (ELISA)
Antibody response to gB and gM antigens were measured by ELISA. The cell lysates of 293 T cells transfected with gB (diluted 1:10) and synthetic peptide representing the highly immunogenic site of gM were used as antigens. Standard ELISA protocols were followed as previously reported.Citation56 One hundred microliters of gB protein (1 µg/ml) or gM peptide (4 µg/ml) diluted in PBS were added to each well. Synthetic peptide representing HCMV gM glycoprotein (sequence: ATASGEEVAV LSHHDSLESR RLREEEDDDD DEDFEDA) (EZBiolab) was dissolved in sterile DMSO to a final concentration of 10 mg/ml and stored at -20°C before use. The endpoint titer was calculated as the serum dilution resulting in absorbance greater than 2 standard deviations above the absorbance in wells incubated with negative control mouse serum.
Microneutralization assay
Microneutralization assay was performed as previously describedCitation49,Citation50. Rabbit sera collected at 1 week after the fourth immunizations were serially diluted in this assay to block the infection of AD169 strain of HCMV to FSK cells. The neutralizing activity for both rabbit sera was complement-dependent s shown in previous protocols.Citation58,Citation59
ELISPOT
To evaluate IFN-gamma producing PBMCs in response to HCMV antigens, cells were stimulated with vaccinia virus recombinant constructs expressing individual HCMV antigens pp65, pp150, IE1, or gB. All detecting monoclonal antibodies applied in this assay were diluted in PBS/0.05% Tween-20. Multiscreen Immobilon P membrane white sterile 96-well plates (IP plates) from Millipore (#S2EM004M99) were coated with 100 µl/well of anti-mouse IFN-gamma at dilution 1:200 (BD Bioscience #18181D) and incubated at 4 °C. After 24 h, the plates were washed 3 times with sterile Dulbecco PBS (Gibco #14190-144) and blocked with 200 μl/well of RPMI 1640 containing 10% fetal calf serum. Fresh splenocytes were added to each well at a concentration of 106 cells in 100 μl volume of complete RPMI medium. Different recombinant vaccinia virus stocks were diluted to a final concentration at 107 pfu/ml in complete RPMI and added in duplicate to cells on the plates. After incubation for 24 h at 37 °C and a washing step (twice with sterile distilled water and 6 times with washing buffer [PBS/0.05% Tween-20]), biotin-labeled rat anti-mouse IFN-gamma antibody was added at dilution 1:400 (BD Biosciences #18112D) and plates were incubated over night at 4 °C. The next day, plates were washed six times with washing buffer and proceeded to the detection step by adding 100 µl/well alkaline phosphatase-streptavidin (BD Bioscience #554065) (diluted 1:800 and incubated at 37 °C for 30 min). Plates were then washed eight times and 100 µl/well of pre-filtered NBT/BCIP substrate (Pierce #34042) was added to each plate. After incubation for 6–8 min at room temp, plates were subsequently washed thoroughly with distilled water and air-dried completely. The number of dark-blue spots was counted under a dissecting microscope. The results were demonstrated as number of IFN-gamma spot-forming cells (SFC) per 106 splenocytes. Antigen-specific T cells were calculated by subtracting the value of negative control wells where cells were infected with the regular vaccinia virus that does not express HCMV antigens.
Statistical analysis
The Student t test (Microsoft Excel software, version 2007) was used to analyze the difference of T cell response results between animal immunization groups measured by IFN-gamma ELISPOT assays. Significance levels were set at a p value less than 0.05.
| Abbreviations: | ||
| CMV | = | cytomegalovirus |
| ELISA | = | enzyme-linked immunosorbent assay |
| ELISPOT | = | enzyme-linked immunosorbent spot assay |
| HBV | = | hepatitis B virus |
| ICS | = | intracellular staining assay |
| IFN | = | interferon |
| VV | = | vaccinia virus |
| VV-WR | = | vaccinia virus - Western Reserve strain |
| MVA | = | modified vaccinia virus Ankara |
Acknowledgments
We are grateful to Dr William Britt (University of Alabama) for providing monoclonal antibody p63-27. We also would like to thank Dr Jill M. Serrano for her careful reading and editing of the manuscript. This study was supported in part by NIH grant AI078073 (SL), R01-AI63356 (DJD). R01-AI103960 (DJD), R01-CA077544 (DJD), P30-CA033572 (City of Hope Comprehensive Cancer Center).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Crough T, Khanna R. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev 2009; 22:76 - 98; http://dx.doi.org/10.1128/CMR.00034-08; PMID: 19136435
- Whitley RJ. Congenital cytomegalovirus infection: epidemiology and treatment. Adv Exp Med Biol 2004; 549:155 - 60; http://dx.doi.org/10.1007/978-1-4419-8993-2_21; PMID: 15250528
- Stratton K, Kurch J, Lawrence R. Vaccine for the 21st Century: a tool for decision making. Washington, D.C.: National Academy Press, 2000.
- Plotkin SA, Starr SE, Friedman HM, Brayman K, Harris S, Jackson S, et al. Effect of Towne live virus vaccine on cytomegalovirus disease after renal transplant. A controlled trial. Ann Intern Med 1991; 114:525 - 31; http://dx.doi.org/10.7326/0003-4819-114-7-525; PMID: 1848053
- Heineman TC, Schleiss M, Bernstein DI, Spaete RR, Yan L, Duke G, et al. A phase 1 study of 4 live, recombinant human cytomegalovirus Towne/Toledo chimeric vaccines. J Infect Dis 2006; 193:1350 - 60; http://dx.doi.org/10.1086/503365; PMID: 16619181
- Pass RF, Zhang C, Evans A, Simpson T, Andrews W, Huang M-L, et al. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med 2009; 360:1191 - 9; http://dx.doi.org/10.1056/NEJMoa0804749; PMID: 19297572
- Abel K, Martinez J, Yue Y, Lacey SF, Wang Z, Strelow L, et al. Vaccine-induced control of viral shedding following rhesus cytomegalovirus challenge in rhesus macaques. J Virol 2011; 85:2878 - 90; http://dx.doi.org/10.1128/JVI.00883-10; PMID: 21191005
- Wussow F, Yue Y, Martinez J, Deere JD, Longmate J, Herrmann A, et al. A vaccine based on the rhesus cytomegalovirus UL128 complex induces broadly neutralizing antibodies in rhesus macaques. J Virol 2013; 87:1322 - 32; http://dx.doi.org/10.1128/JVI.01669-12; PMID: 23152525
- Gibson L, Piccinini G, Lilleri D, Revello MG, Wang Z, Markel S, et al. Human cytomegalovirus proteins pp65 and immediate early protein 1 are common targets for CD8+ T cell responses in children with congenital or postnatal human cytomegalovirus infection. J Immunol 2004; 172:2256 - 64; PMID: 14764694
- Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 2005; 202:673 - 85; http://dx.doi.org/10.1084/jem.20050882; PMID: 16147978
- Berencsi K, Gyulai Z, Gönczöl E, Pincus S, Cox WI, Michelson S, et al. A canarypox vector-expressing cytomegalovirus (CMV) phosphoprotein 65 induces long-lasting cytotoxic T cell responses in human CMV-seronegative subjects. J Infect Dis 2001; 183:1171 - 9; http://dx.doi.org/10.1086/319680; PMID: 11262198
- Bernstein DI, Reap EA, Katen K, Watson A, Smith K, Norberg P, et al. Randomized, double-blind, Phase 1 trial of an alphavirus replicon vaccine for cytomegalovirus in CMV seronegative adult volunteers. Vaccine 2009; 28:484 - 93; http://dx.doi.org/10.1016/j.vaccine.2009.09.135; PMID: 19857446
- Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol 2008; 8:247 - 58; http://dx.doi.org/10.1038/nri2274; PMID: 18323851
- Donnelly JJ, Ulmer JB, Shiver JW, Liu MA. DNA vaccines. Annu Rev Immunol 1997; 15:617 - 48; http://dx.doi.org/10.1146/annurev.immunol.15.1.617; PMID: 9143702
- Ramshaw IA, Ramsay AJ. The prime-boost strategy: exciting prospects for improved vaccination. Immunol Today 2000; 21:163 - 5; http://dx.doi.org/10.1016/S0167-5699(00)01612-1; PMID: 10740236
- Schneider J, Gilbert SC, Hannan CM, Dégano P, Prieur E, Sheu EG, et al. Induction of CD8+ T cells using heterologous prime-boost immunisation strategies. Immunol Rev 1999; 170:29 - 38; http://dx.doi.org/10.1111/j.1600-065X.1999.tb01326.x; PMID: 10566139
- Lu S. Heterologous prime-boost vaccination. Curr Opin Immunol 2009; 21:346 - 51; http://dx.doi.org/10.1016/j.coi.2009.05.016; PMID: 19500964
- Estcourt MJ, Ramsay AJ, Brooks A, Thomson SA, Medveckzy CJ, Ramshaw IA. Prime-boost immunization generates a high frequency, high-avidity CD8(+) cytotoxic T lymphocyte population. Int Immunol 2002; 14:31 - 7; http://dx.doi.org/10.1093/intimm/14.1.31; PMID: 11751749
- McShane H. Prime-boost immunization strategies for infectious diseases. Curr Opin Mol Ther 2002; 4:23 - 7; PMID: 11883691
- Hu SL, Abrams K, Barber GN, Moran P, Zarling JM, Langlois AJ, et al. Protection of macaques against SIV infection by subunit vaccines of SIV envelope glycoprotein gp160. Science 1992; 255:456 - 9; http://dx.doi.org/10.1126/science.1531159; PMID: 1531159
- Harari A, Bart PA, Stöhr W, Tapia G, Garcia M, Medjitna-Rais E, et al. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J Exp Med 2008; 205:63 - 77; http://dx.doi.org/10.1084/jem.20071331; PMID: 18195071
- Dunachie SJ, Walther M, Epstein JE, Keating S, Berthoud T, Andrews L, et al. A DNA prime-modified vaccinia virus ankara boost vaccine encoding thrombospondin-related adhesion protein but not circumsporozoite protein partially protects healthy malaria-naive adults against Plasmodium falciparum sporozoite challenge. Infect Immun 2006; 74:5933 - 42; http://dx.doi.org/10.1128/IAI.00590-06; PMID: 16988273
- Magalhaes I, Sizemore DR, Ahmed RK, Mueller S, Wehlin L, Scanga C, et al. rBCG induces strong antigen-specific T cell responses in rhesus macaques in a prime-boost setting with an adenovirus 35 tuberculosis vaccine vector. PLoS One 2008; 3:e3790; http://dx.doi.org/10.1371/journal.pone.0003790; PMID: 19023426
- Biswas S, Reddy GS, Srinivasan VA, Rangarajan PN. Preexposure efficacy of a novel combination DNA and inactivated rabies virus vaccine. Hum Gene Ther 2001; 12:1917 - 22; http://dx.doi.org/10.1089/104303401753153965; PMID: 11589833
- Xiao-wen H, Shu-han S, Zhen-lin H, Jun L, Lei J, Feng-juan Z, et al. Augmented humoral and cellular immune responses of a hepatitis B DNA vaccine encoding HBsAg by protein boosting. Vaccine 2005; 23:1649 - 56; http://dx.doi.org/10.1016/j.vaccine.2004.10.013; PMID: 15705468
- Spaete RR, Saxena A, Scott PI, Song GJ, Probert WS, Britt WJ, et al. Sequence requirements for proteolytic processing of glycoprotein B of human cytomegalovirus strain Towne. J Virol 1990; 64:2922 - 31; PMID: 2159553
- Wang S, Chou TH, Sakhatskyy PV, Huang S, Lawrence JM, Cao H, et al. Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J Virol 2005; 79:1906 - 10; http://dx.doi.org/10.1128/JVI.79.3.1906-1910.2005; PMID: 15650214
- Lu S, Wyatt R, Richmond JF, Mustafa F, Wang S, Weng J, et al. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/2 and V3 regions. AIDS Res Hum Retroviruses 1998; 14:151 - 5; http://dx.doi.org/10.1089/aid.1998.14.151; PMID: 9462925
- Shen S, Wang S, Britt WJ, Lu S. DNA vaccines expressing glycoprotein complex II antigens gM and gN elicited neutralizing antibodies against multiple human cytomegalovirus (HCMV) isolates. Vaccine 2007; 25:3319 - 27; http://dx.doi.org/10.1016/j.vaccine.2007.01.011; PMID: 17287056
- Gyulai Z, Endresz V, Burian K, Pincus S, Toldy J, Cox WI, et al. Cytotoxic T lymphocyte (CTL) responses to human cytomegalovirus pp65, IE1-Exon4, gB, pp150, and pp28 in healthy individuals: reevaluation of prevalence of IE1-specific CTLs. J Infect Dis 2000; 181:1537 - 46; http://dx.doi.org/10.1086/315445; PMID: 10823751
- Bunde T, Kirchner A, Hoffmeister B, Habedank D, Hetzer R, Cherepnev G, et al. Protection from cytomegalovirus after transplantation is correlated with immediate early 1-specific CD8 T cells. J Exp Med 2005; 201:1031 - 6; http://dx.doi.org/10.1084/jem.20042384; PMID: 15795239
- Britt WJ, Vugler LG, Butfiloski EJ, Stephens EB. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol 1990; 64:1079 - 85; PMID: 2154594
- Shimamura M, Mach M, Britt WJ. Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response. J Virol 2006; 80:4591 - 600; http://dx.doi.org/10.1128/JVI.80.9.4591-4600.2006; PMID: 16611919
- Plotkin SA, Smiley ML, Friedman HM, Starr SE, Fleisher GR, Wlodaver C, et al. Towne-vaccine-induced prevention of cytomegalovirus disease after renal transplants. Lancet 1984; 1:528 - 30; http://dx.doi.org/10.1016/S0140-6736(84)90930-9; PMID: 6142252
- Adler SP, Starr SE, Plotkin SA, Hempfling SH, Buis J, Manning ML, et al. Immunity induced by primary human cytomegalovirus infection protects against secondary infection among women of childbearing age. J Infect Dis 1995; 171:26 - 32; http://dx.doi.org/10.1093/infdis/171.1.26; PMID: 7798679
- Jacobson MA, Sinclair E, Bredt B, Agrillo L, Black D, Epling CL, et al. Antigen-specific T cell responses induced by Towne cytomegalovirus (CMV) vaccine in CMV-seronegative vaccine recipients. J Clin Virol 2006; 35:332 - 7; http://dx.doi.org/10.1016/j.jcv.2005.09.019; PMID: 16387547
- Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, et al. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 2004; 78:10023 - 33; http://dx.doi.org/10.1128/JVI.78.18.10023-10033.2004; PMID: 15331735
- Lilja AE, Shenk T. Efficient replication of rhesus cytomegalovirus variants in multiple rhesus and human cell types. Proc Natl Acad Sci U S A 2008; 105:19950 - 5; http://dx.doi.org/10.1073/pnas.0811063106; PMID: 19064925
- Cui X, Meza BP, Adler SP, McVoy MA. Cytomegalovirus vaccines fail to induce epithelial entry neutralizing antibodies comparable to natural infection. Vaccine 2008; 26:5760 - 6; http://dx.doi.org/10.1016/j.vaccine.2008.07.092; PMID: 18718497
- Liu MA. DNA vaccines: an historical perspective and view to the future. Immunol Rev 2011; 239:62 - 84; http://dx.doi.org/10.1111/j.1600-065X.2010.00980.x; PMID: 21198665
- Schleiss MR. VCL-CB01, an injectable bivalent plasmid DNA vaccine for potential protection against CMV disease and infection. Curr Opin Mol Ther 2009; 11:572 - 8; PMID: 19806506
- Lu S, Wang S, Grimes-Serrano JM. Current progress of DNA vaccine studies in humans. Expert Rev Vaccines 2008; 7:175 - 91; http://dx.doi.org/10.1586/14760584.7.2.175; PMID: 18324888
- Wang S, Kennedy JS, West K, Montefiori DC, Coley S, Lawrence J, et al. Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 2008; 26:3947 - 57; http://dx.doi.org/10.1016/j.vaccine.2007.12.060; PMID: 18724414
- Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, et al, VRC 306 Study Team. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis 2011; 11:916 - 24; http://dx.doi.org/10.1016/S1473-3099(11)70240-7; PMID: 21975270
- Wang S, Parker C, Taaffe J, Solórzano A, García-Sastre A, Lu S. Heterologous HA DNA vaccine prime--inactivated influenza vaccine boost is more effective than using DNA or inactivated vaccine alone in eliciting antibody responses against H1 or H3 serotype influenza viruses. Vaccine 2008; 26:3626 - 33; http://dx.doi.org/10.1016/j.vaccine.2008.04.073; PMID: 18538900
- Suguitan ALJ Jr., Cheng X, Wang W, Wang S, Jin H, Lu S. Influenza H5 hemagglutinin DNA primes the antibody response elicited by the live attenuated influenza A/Vietnam/1203/2004 vaccine in ferrets. PLoS One 2011; 6:e21942; http://dx.doi.org/10.1371/journal.pone.0021942; PMID: 21760928
- Morello CS, Ye M, Spector DH. Development of a vaccine against murine cytomegalovirus (MCMV), consisting of plasmid DNA and formalin-inactivated MCMV, that provides long-term, complete protection against viral replication. J Virol 2002; 76:4822 - 35; http://dx.doi.org/10.1128/JVI.76.10.4822-4835.2002; PMID: 11967299
- Morello CS, Ye M, Hung S, Kelley LA, Spector DH. Systemic priming-boosting immunization with a trivalent plasmid DNA and inactivated murine cytomegalovirus (MCMV) vaccine provides long-term protection against viral replication following systemic or mucosal MCMV challenge. J Virol 2005; 79:159 - 75; http://dx.doi.org/10.1128/JVI.79.1.159-175.2005; PMID: 15596812
- Jacobson MA, Adler SP, Sinclair E, Black D, Smith A, Chu A, et al. A CMV DNA vaccine primes for memory immune responses to live-attenuated CMV (Towne strain). Vaccine 2009; 27:1540 - 8; http://dx.doi.org/10.1016/j.vaccine.2009.01.006; PMID: 19168107
- Kern F, Surel IP, Faulhaber N, Frömmel C, Schneider-Mergener J, Schönemann C, et al. Target structures of the CD8(+)-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J Virol 1999; 73:8179 - 84; PMID: 10482568
- Wang Z, La Rosa C, Maas R, Ly H, Brewer J, Mekhoubad S, et al. Recombinant MVA expressing a soluble form of Glycoprotein B causes durable immunity and neutralizing antibodies against multiple strains of CMV. J Virol 2004; 78:3965 - 76; http://dx.doi.org/10.1128/JVI.78.8.3965-3976.2004; PMID: 15047812
- Wang Z, La Rosa C, Mekhoubad S, Lacey SF, Villacres MC, Markel S, et al. Attenuated poxviruses generate clinically relevant frequencies of CMV-specific T cells. Blood 2004; 104:847 - 56; http://dx.doi.org/10.1182/blood-2003-10-3469; PMID: 15090456
- La Rosa C, Wang Z, Lacey SF, Markel SF, Sharma MC, Martinez J, et al. Characterization of host immunity to cytomegalovirus pp150 (UL32). Hum Immunol 2005; 66:116 - 26; http://dx.doi.org/10.1016/j.humimm.2004.10.008; PMID: 15694996
- Wang Z, La Rosa C, Li Z, Ly H, Krishnan A, Martinez J, et al. Vaccine properties of a novel marker gene-free recombinant modified vaccinia Ankara expressing immunodominant CMV antigens pp65 and IE1. Vaccine 2007; 25:1132 - 41; http://dx.doi.org/10.1016/j.vaccine.2006.09.067; PMID: 17049414
- Lu S, Manning S, Arthos J. Antigen engineering in DNA immunization. Totowa, NJ: Humana Press, 1999:355-74.
- Wang S, Pal R, Mascola JR, Chou TH, Mboudjeka I, Shen S, et al. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology 2006; 350:34 - 47; http://dx.doi.org/10.1016/j.virol.2006.02.032; PMID: 16616287
- Wang S, Joshi S, Lu S. Delivery of DNA to skin by particle bombardment. Methods Mol Biol 2004; 245:185 - 96; PMID: 14707379
- Gonczol E, Furlini G, Ianacone J, Plotkin SA. A rapid microneutralization assay for cytomegalovirus. J Virol Methods 1986; 14:37 - 41; http://dx.doi.org/10.1016/0166-0934(86)90005-4; PMID: 3021795
- Andreoni M, Faircloth M, Vugler L, Britt WJ. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J Virol Methods 1989; 23:157 - 67; http://dx.doi.org/10.1016/0166-0934(89)90129-8; PMID: 2542350
