Abstract
Proliferating Cell Nuclear Antigen (PCNA) is a key protein in DNA replication and repair. The dynamics of replication and repair in live cells is usually studied introducing translational fusions of PCNA. To obviate the need for transfection and bypass the problem of difficult to transfect and/or short lived cells, we have now developed a cell permeable replication and/or repair marker. The design of this marker has three essential molecular components: (1) an optimized artificial PCNA binding peptide; (2) a cell-penetrating peptide, derived from the HIV-1 Trans Activator of Transcription (TAT); (3) an in vivo cleavable linker, linking the two peptides. The resulting construct was taken up by human, hamster and mouse cells within minutes of addition to the media. Inside the cells, the cargo separated from the vector peptide and bound PCNA effectively. Both replication and repair sites could be directly labeled in live cells making it the first in vivo cell permeable peptide marker for these two fundamental cellular processes. Concurrently, we also introduced a quick peptide based PCNA staining method as an alternative to PCNA antibodies for immunofluorescence applications. In summary, we present here a versatile tool to instantaneously label repair and replication processes in fixed and live cells.
Introduction
DNA replication and repair are the most fundamental biological processes in any known organism. Several factors have been evolved to control the fidelity and efficiency of these processes. Deregulation of any these processes can lead to several fatal diseases such as cancer.
Proliferating cell nuclear antigen (PCNA) is a central molecule at the crossroads of DNA replication and DNA repair. Its functions include also other vital epigenetic cellular processes, such as chromatin remodeling, sister-chromatid cohesion, and cell cycle control.Citation1,2 It has been even suggested that PCNA could be the main protein that executes “decisions” made by p53.Citation3 Its importance is strongly highlighted by its structural and sequence homology between distantly evolved organisms such as plants and animals.
Structurally, PCNA forms a homotrimer that encircles DNA forming a sliding clamp () that functions as a loading platform directly interacting in this way with several proteins involved in DNA replication and repair.Citation4 This platform slides along DNA increasing the processivity of DNA polymerase delta during DNA synthesis.Citation5
Figure 1. PCNA marks DNA replication and repair sites. (A) Crystal structure of PCNA (green) encircling the DNA (red) (PDB 1AXC). PCNA serves as a loading platform for several repair and replication factors that bind to a common PCNA region indicated with a dotted line. (B) During the DNA synthesis phase (S-phase) PCNA is recruited to sub-nuclear sites of DNA replication forming distinctive patterns over time that characterize different S-phase stages (shown schematically and with confocal images of GFP-PCNA expressing cells). (C) PCNA associates with sites of DNA damage induced by laser microirradiation. Scale bar 5 μm.
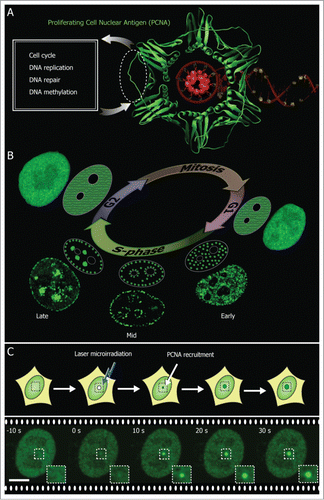
The recruitment of multiple proteins by PCNA must be regulated in a spatio-temporal manner to efficiently synchronize DNA replication and repair. Hundreds of proteins share common PCNA binding sitesCitation6 and it has been proposed that the PCNA binding affinities play a critical regulatory role in the orchestrated progression of DNA repair and replication. It has been shown, for example, that yeast strains expressing mutant forms of PCNA with increased affinity for these proteins exhibit severe alterations in DNA replication and repair. This suggests that intermediate degrees of affinities might have evolved to allow numerous transient interactions in a hierarchical manner.Citation7,8
One important PCNA interaction partner is the tumor suppressor protein p21WAF Citation1 (p21). This protein controls tumor progression by inhibiting cyclin dependent kinases (CDKs) and directly binding to PCNA.Citation9 The C terminus of p21, responsible for its direct binding to PCNA, is well conserved among several PCNA binding partners. However, it is believed that p21 inhibits or disrupts cell cycle progression by having a higher PCNA binding affinity and competitively inhibiting the binding of other PCNA-interacting partners required for DNA replication. This hypothesis has led to the development of several PCNA binding peptides inspired in the C terminus of p21. It has been shown that synthetic peptides derived from p21 can arrest cell cycle progression and even kill cancer cells.Citation10-13
During S phase and DNA damage repair, PCNA is co-localized with sites of active DNA synthesis.Citation14 The development of cell proliferation biomarkers in recent years has provided a much deeper understanding of these processes and has led to new clinical tools. Currently, there are two microscopy-based approaches commonly used to visualize the PCNA in replication and/or repair: (1) immunofluorescence in fixed cells and (2) ectopic expression of fluorescent fusions of PCNA in living cells.
Tumor growth correlates with an increase of PCNA levels. Immunostaining is commonly used to quantify the expression of PCNA in tissue as a diagnostic tool. A potential pitfall in quantifying the levels of PCNA to diagnose tumor growth is that these levels also increase during DNA repair. Therefore, the results of these tests can be easily altered by everyday natural factors such as sun exposure, which leads to UV induced DNA damage and other skin lesions.
Immunostaining methods cannot be performed directly in live cells as it requires cellular fixation and permeabilization. Live cell studies, on the other hand, currently require the transfection of cells with fluorescent fusions of PCNA or other replication factors thereby visualizing the replication foci.Citation14,15 These replication foci form patterns that undergo changes in individual cells throughout S-phase characterizing different stages of DNA synthesis as shown in . If DNA is damaged using laser microirradiation, the repair sites can also be detected by looking at the accumulation of recruited PCNA to the damaged sites as shown in . Successful markers for cell cycle analysis have already made use of these DNA constructs expressing PCNA fluorescent fusion tags.Citation16 A critical limitation of this approach is that cells need to be expressing the fluorescently labeled PCNA fusion protein. This causes a time delay and is not possible or efficient in primary cells or otherwise difficult to transfect cells.
Here we present the design of an instantaneous live cell replication and repair marker. This design is based on three key components: (1) a high affinity PCNA interacting peptide (PIP), (2) a cell-penetrating peptide (CPP) to transport the PIP into the cells, and (3) an intracellular cleavable linker between the two peptides, so that the PIP can get uncoupled from the CPP after reaching the interior of the cells. We first present the rationale of the peptide design. Then we show its uptake in live mammalian cells instantaneously labeling repair and replication sites.
Results
All replication and repair factors seem to bind to a common region in PCNA, more specifically the p21 protein binds at the PCNA inter-domain connecting loop located within the area encircled with dotted lines in . The dual role of PCNA in replication and/or repair was visualized in living cells by transfecting fluorescent fusions of PCNA. In are shown different stages of S phase progression that form distinctive PCNA replication patterns. These characteristic replication patterns might arise as a consequence of higher order chromatin architecture and a timely orchestrated order in which chromatin is replicated. Early replication patterns represent actively transcribed euchromatin, whereas late replication patterns are associated with heterochromatic regions.Citation17 Laser microirradiation produces significant DNA damage leading to high PCNA recruitment at the irradiated sites as shown in . Here, we used these live cell assays for DNA replication and repair, using fluorescent tagged PCNA constructs, to directly test the cell permeability and PCNA binding ability of PIPs.
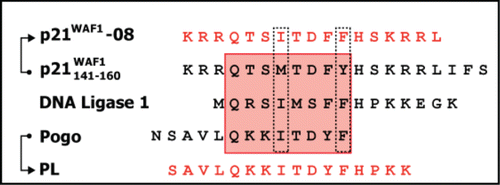
p21 contains a minimal consensus motif critical for its binding to PCNA, also called PIP-box (PCNA interacting protein or peptide), consisting of Q X X (h) X X (a) (a), where Q represents glutamine amino acid, X is any amino acid, (h) a moderately hydrophobic amino acid, and (a) aromatic residues highly hydrophobic side chains.Citation18 The p21141–160 peptide might be evolutionarily optimized to have higher affinity for PCNA than other replication factors. Therefore, we decided to optimize this peptide by searching for a consensus PIP-box motif as shown in . We compared the PIP-box of p21, DNA ligase 1 and Pogo, mutated M147 for I and Y151 for F. The LIFS sequence, p21157–160, is also required to bind with CDK-cyclin complexes.Citation12,19 Since we were interested in targeting exclusively PCNA, to avoid unspecific binding with CDK-cyclin we removed the IFS sequence (p21158–160). The resulting new consensus peptide was named p21–08 (). Using the same set of sequences, p21141–160, DNA ligase I, and Pogo DNA transposase, a peptide called PL (Pogo-Ligase)Citation19,20 was already reported. PL binds to PCNA with an affinity very similar to that of the p21141–160 peptide. The PL peptide was obtained from the sequence of DNA ligase I and Pogo. Therefore, we used those two sequences to optimize instead p21141–157 obtaining p21–08 (). The PL and the newly derived consensus peptide p21–08 are the two PIP sequences we selected for the design of cell-permeable PCNA binding peptides to mark DNA repair and replication sites.
Figure 2. Design of PCNA interacting peptides (PIP). The red box highlights the residues binding to the PCNA binding pocket. The dotted lines indicate the consensus mutations introduced on the p21141–157 peptide to optimize the PIP-box sequence and obtain the p21–08 peptide. The IFS (p21158–160) sequence from p21 was removed to reduce the binding between the p21–08 and the CDK-cyclin and make its binding more selective toward PCNA. Additionally a previously describedCitation19 synthetic peptide PL derived from Pogo and DNA ligase I was used.
First we asked if these peptides would label replication sites in living cells. In are shown the sequences of these PIPs including the wild type p21141–160 peptide and the crystal structure of p21139–160 bound to PCNA (PDB ID 1AXC).Citation21 In we show structures of all these peptides interacting with a PCNA monomer obtained from molecular dynamics simulations. In these simulations we found that the wild type p21141–160, the p21–08, and the PL all got bound to PCNA. More specifically, all these peptides motifs spontaneously bound at distinctive regions of the inter domain connecting loop, PCNA119–133, which is critical for the binding of repair and replication factors. We microinjected these peptides and found that all of them can label PCNA at replication sites in live cells (). This confirmed that the in vitro derived PL peptide and the new p21–08 peptide can bind to PCNA and label replication sites when coupled to a fluorescent tag. Next we asked if any of these labeled peptides would be cell permeable and enter into live cells. We found that when added to the extracellular media, none of these peptides were able to spontaneously cross the cell membrane and label PCNA ().
Figure 3. PCNA interacting peptides bind to PCNA in vivo but are not cell permeable. (A) Crystal structure of the PCNA trimer (green) with the C-terminus region of the p21 protein (red) bound to it (PDB ID 1AXC). The sequences of the p21 peptide and the two highest PCNA binding affinity peptides p21–08 and PL screened are displayed. (B) Structures of the p21 peptides and the two PIPs, i.e., p21–08 and PL sequences obtained from molecular dynamics simulations are shown. (C) Images showing that these peptides ([F-p21], [F-p21–08], [F-PL]) after microinjection clearly co-localize with GFP-PCNA at replication sites. (D) When the peptides are added to the extracellular media they are not able to enter the cells. The abbreviation PC stands for phase contrast and DIC for differential interference contrast. Scale bar 5 μm.
![Figure 3. PCNA interacting peptides bind to PCNA in vivo but are not cell permeable. (A) Crystal structure of the PCNA trimer (green) with the C-terminus region of the p21 protein (red) bound to it (PDB ID 1AXC). The sequences of the p21 peptide and the two highest PCNA binding affinity peptides p21–08 and PL screened are displayed. (B) Structures of the p21 peptides and the two PIPs, i.e., p21–08 and PL sequences obtained from molecular dynamics simulations are shown. (C) Images showing that these peptides ([F-p21], [F-p21–08], [F-PL]) after microinjection clearly co-localize with GFP-PCNA at replication sites. (D) When the peptides are added to the extracellular media they are not able to enter the cells. The abbreviation PC stands for phase contrast and DIC for differential interference contrast. Scale bar 5 μm.](/cms/asset/9858aa8b-b3c8-405a-b46e-a4a86767d2a8/kncl_a_980688_f0003_c.gif)
These peptides need to efficiently cross the cell membrane in order to label replication and repair sites in living cells when added to the extracellular media. It has been shown that cell-penetrating peptides (CPP) efficiently and spontaneously enter live cells transporting other cargo molecules such as peptides.Citation13 Therefore, to make the PIPs membrane permeable we first covalently coupled the p21–08 PIP to a CPP derived from the HIV-1 TAT protein (TAT peptide) able to transduce into live cells and transport other peptides.Citation22-25
The nucleus of a cell expressing fluorescently labeled PCNA was microirradiated, which was accompanied by PCNA recruitment to the damaged site. However, the CPP sequence fused to PIP interfered with the its binding to PCNA () and mistargeted the peptide to the nucleolus, which is a known feature of arginine rich peptides.Citation24,26
Figure 4. Rationale to develop a cell permeable DNA replication and repair marker by targeting PCNA. (A) The nucleus of a cell expressing fluorescently labeled PCNA was microirradiated. PCNA is recruited to damaged DNA sites. Fusion of the PIP with a CPP [F -TAT-p21–08] makes it cell permeable. However, the CPP sequence interferes with the PIP binding to PCNA. (B) Design strategy: the PIP is coupled to a cell-penetrating peptide (in this case the TAT peptide) via a disulfide bridge; after transporting the PIP into the cell the disulfide bridge is reduced; the CPP accumulates mainly in the cytosol and nucleolus, while the PIP is free to reach and bind PCNA. (C) To visualize in live cells the separation of the PIP from CPP, we labeled each sequence with a different fluorescent dye. The TAT peptide labeled with TAMRA is coupled by a disulfide bridge to the PIP peptide labeled with FITC [R-TAT(-SS-)p21–08-F]. It can be seen that the free PIP labels the site of DNA damage, whereas the CPP distributes mainly in the cytosol and nucleolus. Experiments were repeated at least two times. Scale bar 5 μm.
![Figure 4. Rationale to develop a cell permeable DNA replication and repair marker by targeting PCNA. (A) The nucleus of a cell expressing fluorescently labeled PCNA was microirradiated. PCNA is recruited to damaged DNA sites. Fusion of the PIP with a CPP [F -TAT-p21–08] makes it cell permeable. However, the CPP sequence interferes with the PIP binding to PCNA. (B) Design strategy: the PIP is coupled to a cell-penetrating peptide (in this case the TAT peptide) via a disulfide bridge; after transporting the PIP into the cell the disulfide bridge is reduced; the CPP accumulates mainly in the cytosol and nucleolus, while the PIP is free to reach and bind PCNA. (C) To visualize in live cells the separation of the PIP from CPP, we labeled each sequence with a different fluorescent dye. The TAT peptide labeled with TAMRA is coupled by a disulfide bridge to the PIP peptide labeled with FITC [R-TAT(-SS-)p21–08-F]. It can be seen that the free PIP labels the site of DNA damage, whereas the CPP distributes mainly in the cytosol and nucleolus. Experiments were repeated at least two times. Scale bar 5 μm.](/cms/asset/8aef6d58-d711-44cf-be78-3960b55724e0/kncl_a_980688_f0004_c.gif)
This was solved by coupling the PIP to the CPP through a disulfide bridge. This disulfide bridge can be reduced in the reducing environment of the cytosol thereby uncoupling the active peptide (PIP) from the CPP (). To test this coupling design we labeled the CPP with TAMRA and the PIP with FITC. If both peptides get uncoupled in the cytosol, the fluorescent label in each peptide should display a different and distinctive intracellular distribution. Importantly, the uncoupled PIP peptide should be recruited by PCNA. To test this idea we delivered the peptide into laser microirradiated cells (). It can be seen that the PIP peptide accumulated at microirradiated sites, whereas the CPP distributed mainly in the cytosol and nucleolus. This shows that the TAT peptide was able to transport the PIP peptide into the cells, once inside the cells the PIP peptide was uncoupled from the carrier and able to bind PCNA. The uncoupling step seemed to be crucial for the PIP binding to PCNA, since only the decoupled PIP was recruited to the microirradiated site (only the PIP green fluorescent signal increases at the microirradiated sites). Although this PIP (p21–08) was able to label repair sites we did not detect PCNA labeling at replication sites (Fig. S1).
Next, we applied the same CPP coupling design () for the PL sequence. In it can be seen that the PIP fluorescence signal colocalizes with PCNA at laser microirradiated sites in live cells transfected with GFP tagged PCNA. Therefore, the in vivo cleaved PIP marks repair sites.
Figure 5. In vivo immediate labeling of DNA repair sites. Cartoon representation of the experimental protocol and time-lapse snapshots obtained using confocal microscopy are illustrated. GFP tagged PCNA expressing cells were microirradiated. The PL peptide fluorescently labeled and coupled by a disulfide bridge to the TAT peptide [TAT(-SS-)PL-R] was added to the extracellular media. PIP labels PCNA at repair sites. The abbreviation DIC stands for differential interference contrast. Experiments were repeated at least three times. Scale bar 5 μm.
![Figure 5. In vivo immediate labeling of DNA repair sites. Cartoon representation of the experimental protocol and time-lapse snapshots obtained using confocal microscopy are illustrated. GFP tagged PCNA expressing cells were microirradiated. The PL peptide fluorescently labeled and coupled by a disulfide bridge to the TAT peptide [TAT(-SS-)PL-R] was added to the extracellular media. PIP labels PCNA at repair sites. The abbreviation DIC stands for differential interference contrast. Experiments were repeated at least three times. Scale bar 5 μm.](/cms/asset/7041264a-568c-4e0b-a0f1-500d37a30e81/kncl_a_980688_f0005_c.gif)
This peptide was also able to label DNA replication sites. In it is shown the nucleus of cells expressing GFP tagged PCNA along different stages of the cell cycle. The PIP added to the extracellular media was able to reach the interior of living cells, co-localize with PCNA at replication sites, and distinctively label replicating cells at different S-phase stages. At higher concentrations though, the peptide inhibited DNA synthesis as measured by detecting nucleotide incorporation (Fig. S2).
Figure 6. In vivo immediate labeling of DNA replication sites in S-phase. Cartoon representation of the experimental protocol and time-lapse snapshots obtained using confocal microscopy are illustrated. GFP tagged PCNA expressing cells at different cell cycle stages recognizable by the PCNA sub-nuclear patterns were identified and the same peptide as in , PL coupled to TAT peptide via disulfide bridge [TAT(-SS-)PL-R], when added to the extracellular media, labeled the sites of active DNA replication. Experiments were repeated at least three times. Scale bar 5 μm.
![Figure 6. In vivo immediate labeling of DNA replication sites in S-phase. Cartoon representation of the experimental protocol and time-lapse snapshots obtained using confocal microscopy are illustrated. GFP tagged PCNA expressing cells at different cell cycle stages recognizable by the PCNA sub-nuclear patterns were identified and the same peptide as in Figure 5, PL coupled to TAT peptide via disulfide bridge [TAT(-SS-)PL-R], when added to the extracellular media, labeled the sites of active DNA replication. Experiments were repeated at least three times. Scale bar 5 μm.](/cms/asset/8308c3d7-bb49-4765-8207-f03cfd277c8c/kncl_a_980688_f0006_c.gif)
The above results also suggest that these peptides could potentially be used in the place of antibodies in fixed cells, which can be particularly useful for common immunostaining applications. In it is shown that the PL alone can be used in fixed and membrane permeabilized cells to label ectopically expressed () and endogenous PCNA ().
Figure 7. PCNA interacting peptide (PIP) labels replication and repair in fixed cells. (A) Here replication and repair sites can be directly visualized in fixed cells without the need of antibodies by PL peptide [F-PL] in the presence of ectopically expressed PCNA. (B) The same PL peptide clearly marks the endogenous PCNA. Experiments were repeated at least three times. Scale bar 5 μm.
![Figure 7. PCNA interacting peptide (PIP) labels replication and repair in fixed cells. (A) Here replication and repair sites can be directly visualized in fixed cells without the need of antibodies by PL peptide [F-PL] in the presence of ectopically expressed PCNA. (B) The same PL peptide clearly marks the endogenous PCNA. Experiments were repeated at least three times. Scale bar 5 μm.](/cms/asset/44b7ac40-a81b-4615-a048-64e0ad7ecece/kncl_a_980688_f0007_c.gif)
The cell-permeable PIP (Fig. S3), on the other hand, after fixation still marks repair sites but in the case of replication it got redistributed and accumulated in the nucleolus. This indicates that live cells labeled with the cell permeable PIP can be fixed for further postmortem analysis only for DNA repair without disrupting the PCNA labeling by PIP.
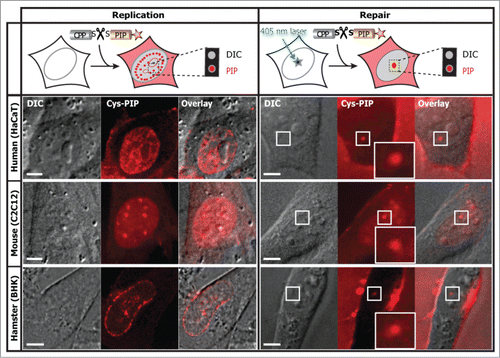
Finally, in it is shown that the cell permeable PIP, obtained by coupling the TAT and the PL peptide through the intracellular cleavable disulfide bridge, also labeled DNA replication and repair sites in live cells from different species.
Figure 8. The cell permeable and intracellular cleavable PIP directly labels DNA replication and repair in different cell types and species. The PL peptide fluorescently labeled and coupled by a disulfide bridge to the TAT peptide was added to the extracellular media on a variety of cells as indicated, with and without laser microirradiation. In the absence of labeled PCNA the PIP allows a clear classification of S-phase replication stages and damaged DNA sites. The abbreviation DIC stands for differential interference contrast. Experiments were repeated at least three times. Scale bar 5 μm.
Discussion
In this study, we designed a cell permeable peptide able to label DNA replication and repair sites in live cells. The strategy was essentially based on coupling a fluorescent labeled high affinity PCNA binder peptide (PIP) to a cell-penetrating peptide.
When the PIPs were microinjected into the cells these peptides were able to label PCNA. However, none of them were able to spontaneously cross into the cells when added to the extracellular media. To overcome this limitation the PIPs were coupled to a cell-penetrating peptide. When the PIP and the cell-penetrating peptide were covalently coupled we observed that the peptides were able to cross the plasma membrane but the binding and labeling of PCNA was inhibited. At least partially, the reason for this could be that the TAT peptide sequence miss-localized the PIPs. The TAT peptide localizes mainly in the cytosol and nucleolusCitation26 while the PIPs need to get distributed in the nucleoplasm to reach and bind to PCNA effectively.
To this end we coupled the TAT and the PIPs via a disulfide bridge that displayed intracellular cleavage. This partially restored the PCNA binding of the p21–08 PIP. This peptide was able to translocate into the cells and label DNA damaged sites but was unable to label replication sites. It is possible that this is a consequence of the mutation introduced in the p21–08 original sequence by the addition of the cysteine amino acid. The dye linker in this case was also different from the microinjected version of the peptide. All these small changes could have potentially altered the p21–08 PIP's binding affinity to PCNA. Although, the reason why the cell permeable disulfide coupled p21–08 peptide was able to label repair sites and not replicating sites requires further research, this could still be potentially very advantageous to selectively label repair sites.
When the same design strategy was used for the PL peptide, we found that this newly engineered peptide was cell permeable and able to instantaneously label both DNA replication and repair sites in live cells.
We observed that the PCNA labeling is transient, indicating that the PIPs are being actively degraded in live cells. Depending on the particular application, there are several strategies that could be further adapted to avoid degradation and extend the intracellular stability of these peptides, such as the introduction of unnatural amino acids or by using PEGylated peptides as previously described.Citation27 However, the best approach is not obvious, since as shown in the case of p21–08 even small modifications in the sequence can significantly alter the PCNA binding of these peptides. All peptides in this study contain also the “PIP degron” motif (Q XX (h) TD (a)(a) XXX K/R).Citation28 It has been shown that although this motif enhances PCNA binding the last amino acid (K or R) is critical for degradation. This degradation seems to play a regulatory role to avoid proteins bearing this motif from blocking DNA repair.Citation29 Therefore, mutating this last amino acid offers a simple alternative route to enhance stability of these peptides potentially leading to a simultaneous increase in labeling and blocking DNA repair.
We have shown also that labeled PL, uncoupled to the TAT peptide, can be used on fixed cells following standard fixation protocols. Therefore, this peptide could also be used as a quick PCNA marker replacing primary and/or secondary antibodies used in immunofluorescence staining methods.
Peptides with high PCNA binding affinity derived from consensus motifs, containing the PIP box motifCitation19 (such as the peptides presented here) or alternatively peptides containing the APIM consensus motif,Citation30 possess the inherent ability to competitively interfere with the PCNA recruitment of replication and repair factors. Covalently coupling these peptides to cell-penetrating peptides shows great potential as an effective route to transport these peptides and inhibit proliferative processes (see also Fig. S2).Citation13,31 In particular, it has been reported that both the PL and p21 peptides interfere with DNA replication.Citation13,19 The results presented here outline a general strategy to design and efficiently deliver these functional peptides using cell-penetrating peptides providing a versatile tool to potentially target proliferative diseases. Consequently, the cell permeable PCNA binding peptides presented here offer interesting therapeutic venues to selectively target and distinguish replication and repair.
In summary, here we have designed the first cell permeable PCNA binding peptide that instantaneously labels cells undergoing DNA repair and replication across mammalian species.
Materials and Methods
Peptides
Peptides used for this study () were synthesized by Biosyntan GmbH (Berlin, Germany). TAMRA-5-tetramethylrhodamine and FITC-fluorescein-isothiocyanate were the fluorophores used for labeling the peptides. Peptide stock solutions were prepared by dissolving the lyophilized peptides in deionized autoclaved water.
Table 1. Summary of peptides
Plasmids
Mammalian expression constructs encoding translational fusions of GFP-PCNA, RFP-PCNA were described in references 32 and 33. pFRT-B-OPCNA was generated from pEF5/FRT/V5 Topo (Invitrogen) by replacing hygromycin with blasticidin and inserting Orange-PCNA after the EF1 α promoter.
Cells, culture, transfection, and microinjection
Human cell line HeLa was cultured in Dulbecco's modified eagle medium (DMEM) and HaCaT with Roswell Park Memorial Institute (RPMI) 1640 medium, both supplemented with 10% fetal calf serum, 50 μg/ml gentamicin, and 2 mM glutamine. Mouse myoblasts C2C12Citation34 cells were cultured in DMEM supplemented with 20% fetal calf serum, 50 μg /ml gentamicin, and 2 mM glutamine. Mouse 3T3 fibroblasts stably expressing Orange tagged PCNA were generated by Flp recombinase mediated site specific integration of pFRT-B-OPCNA in Flp-In-3T3TM cells (Invitrogen) according to the manufacturer's instructions. Stable clones were selected with blasticidin 2.5 μg/ml in 10% fetal calf serum, 50 μg/ml gentamicin, and 2 mM glutamine. Baby hamster kidney cell line was grown in Glasgow minimal essential medium supplemented with 10% fetal calf serum, 50 μg/ml gentamicin, 2 mM glutamine, and 150 μg/ml hygromycin B as previously described.Citation35
All cell culture media was purchased from Sigma-Aldrich. All the cell lines were grown at 37 °C in a humidified atmosphere with 5% CO2. For transfecting the cells polyethylenimine was used as previously described.Citation36
Microinjection of the peptides was performed using an Eppendorf microinjection and micromanipulation system mounted on a Zeiss LSM510Meta confocal microscope. All the peptides were diluted to 100 μM in PBS and injected into the cells. The parameters for cytoplasmic microinjection are two seconds injection with 50 fPa injection pressure and a constant back pressure of 20 fPa, i.e., 30 fPa final injection pressure.
Fluorescent labeling with PIP
Cells were fixed with 3.7% formaldehyde for seven minutes at room temperature. They were permeabilized with 0.5% Triton X-100 for 15 min and washed with PBS. Fluorescently labeled PL peptide was diluted to 13 μM final concentration with PBS, added to the fixed cells, incubated for 30 min at room temperature and washed with ice cold PBS gently. Hence replication and repair sites can be directly visualized in fixed cells without the need of PCNA antibodies. In case of staining for multiple epitopes, normal immunostaining procedure was followed with other antibodies and then the peptide PIP was added followed by 5 min post-fixation with 3.7% formaldehyde. The 4',6-diamidino-2-phenylindole (DAPI) was used to stain the DNA content. The coverslips were mounted in Mowiol 4–88 (Sigma-Aldrich Chemie). PIP binding is not stable to extensive washes when used in combination with other antibody stainings. It should be added at the end avoiding further washes.
In situ replication assay
To assay for DNA synthesis inhibition by the peptides, the thymidine analog BrdU was added to the cells at 100 μM final concentration for 20 min. BrdU incorporation during DNA replication was visualized as described,Citation33 using a rat monoclonal anti BrdU antibody (Gentaur catalog no. OBT0030CX; 1:100 dilution).
Molecular dynamics simulations
Molecular dynamics simulations were performed using the GROMACS package, version 4.5.6,Citation37 and the Amber ff99SBCitation38 force field. The temperature of the system was kept constant, coupling independently each group of molecules at 300 K with a v-rescale thermostat. The pressure was coupled to a Berendsen barostat at 1 atm isotropically. The temperature and pressure time constants of the coupling were 0.2 and 2 ps, respectively. The integration of the equations of motion was performed by using a leap frog algorithm with a time step of 2 fs. Periodic boundary conditions were implemented in all systems. A cutoff of 1 nm was implemented for the Lennard-Jones interactions and the direct space part of the Ewald sum for Coulombic interactions. The Fourier space part of the Ewald splitting was computed by using the particle-mesh Ewald method, with a grid length of 0.11 nm and using a cubic spline interpolation. The systems were composed of a PCNA monomer, one peptide, 15 000 water molecules, and potassium ions to neutralize the total electric charge of each system. The PCNA crystal structure conformation 1VYMCitation19 from the protein data bank was used as the initial conformation of the PCNA monomer. The PCNA monomer was placed at the center of a cubic periodically replicated simulation box and the peptides were placed in a stretched conformation at a corner of each box. The total simulation time for each system expanded 2 μs.
Live cell peptide uptake, microscopy, and microirradiation
Peptides were further diluted in DMEM medium (SIGMA-Aldrich, Steinem, Germany) to a final concentration of 15 μM and directly added to the cells. To visualize the DNA replication, cells expressing fluorescently tagged PCNA or not were directly imaged. Whereas for visualizing the DNA repair sites, damage was induced by selecting specific spots in the nucleus of the cells and microirradiating with a 405 nm laser as previously described.Citation39 After the laser induced DNA damage, peptides at a concentration of 13 μM were added to the cells and the recruitment of the peptide at the repair site was monitored. Imaging for longer time points (>10 min) requires removal of the peptides followed by the addition of growth media to prevent excess uptake by the cells, and was preferentially adopted for repair experiments. Confocal microscopic images were collected using an Ultra VIEW VoX spinning disc system (Perkin Elmer) or on a Nikon Ti microscope equipped with an oil immersion Plan-Apochromat × 60/1.45 NA objective lens (pixel size in XY = 111 nm, Z-step = 0.3–1 μm) with laser lines at 405 nm, 488 nm, and 561 nm. During this time cells were kept in closed chamber designed to fit the microscopy stage at 37 °C in a humidified atmosphere with 5% CO2 using the ACU control system from Olympus.
Microirradiation experiments were performed with 405 nm diode laser set to maximum power at 100% transmission available in the Nikon Ti spinning disc microscope. Preselected spots of 1 μm in diameter within the nucleus were microirradiated for 1.2 s. Prebleach and post bleach images were recorded based on the individual peptide recruitment kinetics.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We thank Petra Domaing and Anne Lehmkuhl for excellent technical assistance. We are indebted to Roger Y Tsien for the gift of mRFP and mOrange cDNAs.
Supplemental Data
Download PDF (2 MB)Funding
This work was supported by grants of the German Research Council (DFG GRK 1657 and DFG CA198/7) and the National Institutes of Health (NIH R01-GM086801). This work used the Extreme Science and Engineering Discovery Environment (XSEDE), request number: MCB130086 and MCB140075.
Supplemental Materials
Supplemental materials may be found on the publisher's website: http://www.tandfonline.com/kncl
References
- Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci 2003; 116:3051-60; PMID:12829735; http://dx.doi.org/10.1242/jcs.00653
- Görisch SM, Sporbert A, Stear JH, Grunewald I, Nowak D, Warbrick E, Leonhardt H, Cardoso MC. Uncoupling the replication machinery: replication fork progression in the absence of processive DNA synthesis. Cell Cycle 2008; 7:1983-90; PMID:18604168; http://dx.doi.org/10.4161/cc.7.13.6094
- Paunesku T, Mittal S, Protić M, Oryhon J, Korolev SV, Joachimiak A, Woloschak GE. Proliferating cell nuclear antigen (PCNA): ringmaster of the genome. Int J Radiat Biol 2001; 77:1007-21; PMID:11682006; http://dx.doi.org/10.1080/09553000110069335
- Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM 3rd. XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res 2004; 32:2193-201; PMID:15107487; http://dx.doi.org/10.1093/nar/gkh556
- Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature 1987; 326:515-7; PMID:2882423; http://dx.doi.org/10.1038/326515a0
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell 2007; 129:665-79; PMID:17512402; http://dx.doi.org/10.1016/j.cell.2007.05.003
- Fridman Y, Palgi N, Dovrat D, Ben-Aroya S, Hieter P, Aharoni A. Subtle alterations in PCNA-partner interactions severely impair DNA replication and repair. PLoS Biol 2010; 8:e1000507; PMID:20967232; http://dx.doi.org/10.1371/journal.pbio.1000507
- Mailand N, Gibbs-Seymour I, Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol 2013; 14:269-82; PMID:23594953; http://dx.doi.org/10.1038/nrm3562
- Trimis G, Chatzistamou I, Politi K, Kiaris H, Papavassiliou AG. Expression of p21waf1/Cip1 in stromal fibroblasts of primary breast tumors. Hum Mol Genet 2008; 17:3596-600; PMID:18713757; http://dx.doi.org/10.1093/hmg/ddn252
- Ball KL, Lain S, Fâhraeus R, Smythe C, Lane DP. Cell-cycle arrest and inhibition of Cdk4 activity by small peptides based on the carboxy-terminal domain of p21WAF1. Curr Biol 1997; 7:71-80; PMID:8999999; http://dx.doi.org/10.1016/S0960-9822(06)00029-7
- Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol 1995; 5:275-82; PMID:7780738; http://dx.doi.org/10.1016/S0960-9822(95)00058-3
- Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther 2002; 1:639-49; PMID:12479224
- Tünnemann G, Martin RM, Haupt S, Patsch C, Edenhofer F, Cardoso MC. Cargo-dependent mode of uptake and bioavailability of TAT-containing proteins and peptides in living cells. FASEB J 2006; 20:1775-84; PMID:16940149; http://dx.doi.org/10.1096/fj.05-5523com
- Cardoso MC, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell 1993; 74:979-92; PMID:8402887; http://dx.doi.org/10.1016/0092-8674(93)90721-2
- Cardoso MC, Joseph C, Rahn HP, Reusch R, Nadal-Ginard B, Leonhardt H. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J Cell Biol 1997; 139:579-87; PMID:9348276; http://dx.doi.org/10.1083/jcb.139.3.579
- Easwaran HP, Leonhardt H, Cardoso MC. Cell cycle markers for live cell analyses. Cell Cycle 2005; 4:453-5; PMID:15701967; http://dx.doi.org/10.4161/cc.4.3.1525
- Chagin VO, Stear JH, Cardoso MC. Organization of DNA replication. Cold Spring Harb Perspect Biol 2010; 2:a000737; PMID:20452942; http://dx.doi.org/10.1101/cshperspect.a000737
- Warbrick E, Heatherington W, Lane DP, Glover DM. PCNA binding proteins in Drosophila melanogaster : the analysis of a conserved PCNA binding domain. Nucleic Acids Res 1998; 26:3925-32; PMID:9705499; http://dx.doi.org/10.1093/nar/26.17.3925
- Kontopidis G, Wu SY, Zheleva DI, Taylor P, McInnes C, Lane DP, Fischer PM, Walkinshaw MD. Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc Natl Acad Sci U S A 2005; 102:1871-6; PMID:15681588; http://dx.doi.org/10.1073/pnas.0406540102
- Zheleva DI, Zhelev NZ, Fischer PM, Duff SV, Warbrick E, Blake DG, Lane DP. A quantitative study of the in vitro binding of the C-terminal domain of p21 to PCNA: affinity, stoichiometry, and thermodynamics. Biochemistry 2000; 39:7388-97; PMID:10858286; http://dx.doi.org/10.1021/bi992498r
- Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 1996; 87:297-306; PMID:8861913; http://dx.doi.org/10.1016/S0092-8674(00)81347-1
- Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988; 55:1189-93; PMID:2849510; http://dx.doi.org/10.1016/0092-8674(88)90263-2
- Herce HD, Garcia AE. Cell penetrating peptides: how do they do it? J Biol Phys 2007; 33:345-56; PMID:19669523; http://dx.doi.org/10.1007/s10867-008-9074-3
- Lättig-Tünnemann G, Prinz M, Hoffmann D, Behlke J, Palm-Apergi C, Morano I, Herce HD, Cardoso MC. Backbone rigidity and static presentation of guanidinium groups increases cellular uptake of arginine-rich cell-penetrating peptides. Nat Commun 2011; 2:453; PMID:21878907; http://dx.doi.org/10.1038/ncomms1459
- Herce HD, Deng W, Helma J, Leonhardt H, Cardoso MC. Visualization and targeted disruption of protein interactions in living cells. Nat Commun 2013; 4:2660; PMID:24154492; http://dx.doi.org/10.1038/ncomms3660
- Martin RM, Tünnemann G, Leonhardt H, Cardoso MC. Nucleolar marker for living cells. Histochem Cell Biol 2007; 127:243-51; PMID:17205309; http://dx.doi.org/10.1007/s00418-006-0256-4
- Nischan N, Chakrabarti A, Serwa RA, Bovee-Geurts PH, Brock R, Hackenberger CP. Stabilization of peptides for intracellular applications by phosphoramidate-linked polyethylene glycol chains. Angew Chem Int Ed Engl 2013; 52:11920-4; PMID:24039043; http://dx.doi.org/10.1002/anie.201303467
- Havens CG, Walter JC. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev 2011; 25:1568-82; PMID:21828267; http://dx.doi.org/10.1101/gad.2068611
- Tsanov N, Kermi C, Coulombe P, Van der Laan S, Hodroj D, Maiorano D. PIP degron proteins, substrates of CRL4Cdt2, and not PIP boxes, interfere with DNA polymerase η and κ focus formation on UV damage. Nucleic Acids Res 2014; 42:3692-706; PMID:24423875; http://dx.doi.org/10.1093/nar/gkt1400
- Gilljam KM, Feyzi E, Aas PA, Sousa MM, Müller R, Vågbø CB, Catterall TC, Liabakk NB, Slupphaug G, Drabløs F, et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. J Cell Biol 2009; 186:645-54; PMID:19736315; http://dx.doi.org/10.1083/jcb.200903138
- Müller R, Misund K, Holien T, Bachke S, Gilljam KM, Våtsveen TK, Rø TB, Bellacchio E, Sundan A, Otterlei M. Targeting proliferating cell nuclear antigen and its protein interactions induces apoptosis in multiple myeloma cells. PLoS One 2013; 8:e70430; PMID:23936203; http://dx.doi.org/10.1371/journal.pone.0070430
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J Cell Biol 2000; 149:271-80; PMID:10769021; http://dx.doi.org/10.1083/jcb.149.2.271
- Sporbert A, Domaing P, Leonhardt H, Cardoso MC. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res 2005; 33:3521-8; PMID:15972794; http://dx.doi.org/10.1093/nar/gki665
- Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 1977; 270:725-7; PMID:563524; http://dx.doi.org/10.1038/270725a0
- Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat Cell Biol 2000; 2:871-8; PMID:11146650; http://dx.doi.org/10.1038/35046510
- Casas-Delucchi CS, Becker A, Bolius JJ, Cardoso MC. Targeted manipulation of heterochromatin rescues MeCP2 Rett mutants and re-establishes higher order chromatin organization. Nucleic Acids Res 2012; 40:e176; PMID:22923521; http://dx.doi.org/10.1093/nar/gks784
- Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013; 29:845-54; PMID:23407358; http://dx.doi.org/10.1093/bioinformatics/btt055
- Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010; 78:1950-8; PMID:20408171
- Mortusewicz O, Leonhardt H. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol Biol 2007; 8:81; PMID:17880707; http://dx.doi.org/10.1186/1471-2199-8-81

