Abstract
As a transcription factor, Yin Yang 1 (YY1) regulates the transcription of a dazzling list of genes and the number of its targets still mounts. Recent studies revealed that YY1 possesses functions independent of its DNA binding activity and its regulatory role in tumorigenesis has started to emerge.
YY1 was discovered as a transcription factor and its transcriptional activity can be converted from a repressor to an activator by viral oncoprotein E1A.Citation1 As an ubiquitously expressed transcription factor, YY1 has been demonstrated to regulate cell proliferation and differentiation, and its deficiency in mice results in peri-implantation lethality during embryonic development.Citation2 However, YY1 has attracted the interest of researchers not only because of its essential role in normal cell growth, but also due to its aberrant expression and potential regulatory function in different cancers. YY1 has been reported to modulate a mounting list of genes, many of which are key players in different signaling pathways regulating cancer development and progression, such as c-myc, c-fos, ERBB2, E1A and p53.Citation3,Citation4 Meanwhile, YY1 also physically interacts with a number of proteins regulating cell proliferation and apoptosis, such as p53, Mdm2, Ezh2, Rb, caspases and HDACs.Citation3 In addition, YY1 gene expression can be stimulated by several growth factors, while antiproliferative signals tend to antagonize its expression.Citation3 Therefore, as a multifunctional mediator of different signaling pathways, YY1 potentially acts as a critical regulator in cancer development and progression and likely plays a proliferative or oncogenic role in these processes.
YY1: Beyond a Transcription Factor
To date, most published data have demonstrated the activities of YY1 as a transcription factor. YY1 can either activate or repress gene expression, depending on the cofactors that it recruits. The promoters of these genes normally contain at least one of the two most frequent core binding elements of YY1: CCAT and ACAT.Citation5 YY1 plays an essential role in embryonic development.Citation2 Consistently, many YY1-targeted genes are key regulators of cell growth and differentiation, such as several mitochondrial proteins, Cdc6 and cyclin D1.Citation3 A major portion of these targets are key components of signaling pathways related to apoptosis and oncogenic transformation.Citation3,Citation4
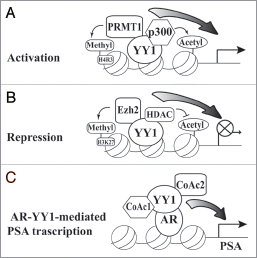
Despite the well-established regulation of YY1 as a transcription factor, several reports, including ours, have demonstrated the role of YY1 as a transcription cofactor and its activity independent of its DNA binding. In our recent study, we observed YY1-dependent expression of prostatespecific antigen (PSA) in prostate cancer cells.Citation6 Due to the well-recognized function of YY1 as a transcription factor, we first tested whether YY1 directly regulates PSA transcription. When we mutated the only YY1 binding element in the PSA promoter, YY1-mediated transcription of PSA promoter was unaffected. Since androgen receptor (AR) is a well-studied transcription activator of PSA, we therefore asked whether YY1 is involved in AR-mediated transcription of PSA. Following this path, we discovered that YY1 directly interacts with AR and enhances the association of AR with an androgen response element (ARE). Importantly, we observed that a YY1 mutant deficient of binding to AR lost the stimulating effect on the PSA promoter that wild type YY1 has. Hence, these studies suggest that YY1-AR interaction, but not YY1-DNA association, is essential to YY1-activated PSA expression. Therefore, instead of directly acting as a transcription factor, YY1 plays a role of a cofactor and its DNA binding activity is dispensable in mediating PSA expression. It is noteworthy that AR interacts with the C-terminal of YY1,Citation6 where its DNA binding domain, the zinc finger region, is located. Therefore, it is unlikely that YY1 molecule can associate with its DNA binding element and AR protein concurrently. The domain mapping studies indicate that the N-terminal of YY1 possesses transcriptional activation activity, and the N-terminal and middle region recruit many proteins with chromatin remodeling functions.Citation7,Citation8 Therefore, with the C-terminal binding to AR, the N-terminal of YY1 will likely recruit other coactivators and consequently promote PSA gene transcription.
This is not the first observation of YY1 exhibiting activity independent of its DNA binding affinity. Our previous study also indicated that YY1 enhances the Hoxa11-DNA association and is required for the recruitment of HDAC2 by Hoxa11.Citation9 Moreover, we and others reported that YY1 stimulates Mdm2-mediated p53 ubiquitination and degradation.Citation10–Citation12 In this study, both wild-type YY1 and its DNA-binding deficient mutant promoted p53 ubiquitination. Importantly, this effect of YY1 on p53 could be visualized in a reconstituted ubiquitination system in vitro.Citation10 Additionally, the observation of altered translocation between nuclear and cytoplasm at different cell cycle stages also suggests that YY1 is involved in biological processes in addition to mediating gene transcription.Citation13,Citation14
The full length of YY1 protein has never been crystallized for structure analysis, except for the cocrystal study of its zinc finger region with the adeno-associated virus P5 promoter.Citation15 Therefore, the actual three-dimensional structure of YY1 activation domains at its N-terminal and middle region remains unclear. However, the amino acid composition of YY1 reveals some of its potential structural features. YY1 protein has two acidic regions at the N-terminal. Especially, an elevenrun of glutamic/asparatic acids is present at residues 43–53 and an eleven-run of histidines consists of residues 70–80 of human YY1, which are both wellconserved in its orthologs from rat, mouse and chick. The electronic charge of histidines may be altered with slight pH changes in the microenvironment. Therefore, the N-terminal of YY1 may employ electrostatic force to interact with different proteins, especially those with positive charges, such as histones. Between residues 150 to 200, there are four clusters of three- to five-runs of glycines that provide large flexibility to this region, which interacts with several proteins, including p300, HDACs, c-Myc, p53 and p14ARF (reviewed in refs. Citation3 and Citation7). Overall, these features may determine the potential of YY1 as a scaffold protein that recruits other transcription regulators to mediate gene expression when its C-terminal binds to either DNA or other transcription factors.
The discovery of YY1 as a cofactor of AR extended our understanding of YY1-mediated transcription. YY1 interacts with a number of protein modifiers that mediate various posttranslational modifications. Early studies by Shi and Seto demonstrated the association of YY1 with p300 and HDACs, regulating histone acetylation and deacetylation, respectively.Citation16,Citation17 Studies by Sartorelli and Seto groups also revealed that YY1 recruits Ezh2 and PRMT1 to mediate histone methylation on lysine and arginine residues, respectively.Citation18,Citation19 In addition, our recent studies demonstrated the association of YY1 with Mdm2, PIASy and Ubc9, which regulate protein ubiquitination and sumoylation, respectively.Citation10,Citation20 Therefore, when binding to its own responsive element or recruited by another transcription factor, YY1 can likely provide a platform for the assembly of a scaffold with different transcriptional machineries where many other cofactors are recruited and assembled together. Especially, when a particular protein modifier binds to YY1, posttranslational modifications may occur to both histones and other recruited cofactors on a target promoter to modulate their functions. This interplay between the recruitment and modification will therefore determine the expression status of the regulated genes. Generally, p300-mediated histone acetylation and PRMT1-mediated histone H4-R3 methylation cause gene activation, while histone deacetylation by HDACs and H3-K27 tri-methylation by Ezh2 silence target genes ( and B). In addition, Mdm2 can cause histone ubiquitination and establishes transcriptional repression.Citation21 Most transcription factors, such as AR,Citation22 p53,Citation23 and YY1 itself,Citation3 also undergo various modifications, which differentially regulate their transcriptional activities. When YY1 acts as a transcription coactivator in AR-mediated PSA gene expression, it is likely that YY1 recruits other coactivators that are yet to be identified to promote transcription (). It is noteworthy that, as a cofactor, the cellular levels of YY1 may determine the expression status of a target gene. Therefore, while a medium increase of YY1 stimulates AR-mediated transcription, further increases of YY1 can compromise this activation.Citation6 One possible reason for this phenomenon is that excessive YY1 associates with other coactivators to be recruited and therefore interferes with the transcriptional activation of a target gene, the so-called “squelching effect”.Citation24
YY1: Involvement in Prostate Cancer
YY1 has been suggested as a regulatory protein, a potential therapeutic target and a prognostic marker of cancers based on its aberrant expression in tumors and the properties of its target genes and interacting proteins.Citation3,Citation25 YY1 overexpression has been detected in multiple cancers, including prostate cancer.Citation26 Bonavida group made a number of seminal discoveries that link YY1 to prostate cancer development and therapy.Citation4 They demonstrated a significant association between YY1 activity and the expression of both cytokine and death receptor. Mechanistically, YY1 negatively regulates the expression of both Fas and the death receptor 5 (DR5), and consequently endows prostate cancer cells with resistance to Fas-induced or TRAIL-induced apoptosis. Consistently, in a tissue microarray study, they observed that elevated YY1 expression correlates with higher morphologic grades or malignant histological phenotypes in prostate cancer samples from 246 patients.Citation27 Our finding of YY1 as a coactivator of AR in promoting PSA transcription also implicates that YY1 potentially regulates prostate cancer development and progression through stimulating AR function. Especially, in the absence of androgen, we observed a modest activation by YY1 on AR-dependent transcription of the PSA promoter,Citation6 suggesting that, as a coactivator, overexpressed YY1 may be an essential regulator to the AR-signaling pathway in androgen independent prostate cancer.
Prostate cancer is a major public health problem among men of many countries. While locally confined prostate tumors can be treated by surgery and radiation therapy, advanced and relapsed prostate cancers are primarily handled by inhibiting the androgen signaling pathway. This can be typically achieved by androgen deprivation therapy including castration and administration of androgen antagonists. However, despite these therapies of androgen withdrawal, prostate cancer unavoidably progresses to the androgen-independent state. Even at low androgen levels, androgen-responsive genes in these tumors restore their expression nearly to the pre-treatment levels.Citation28 This strongly suggests that AR still plays an essential role in the growth and survival of recurred or advanced prostate cancers, even in androgen-deprived conditions. In fact, such prostate cancers retain high expression levels of AR and are still AR-dependent.Citation29 Therefore, AR is still a valid target for the therapeutic intervention of androgenindependent prostate cancers.
The aberrantly activated AR-signaling pathway in prostate cancer can be caused by different deregulations, including AR overexpression, AR mutation, increased sensitivity of AR to low androgen levels and ligand-independent AR activation.Citation30 Several mechanisms may lead to enhanced AR activity or sensitivity, including posttranslational modifications and stimulation of overexpressed coactivators.Citation30 Our observation on that overexpressed YY1 enhances the transcriptional activity of AR suggests that YY1 and its interaction with AR or other transcription cofactors can be potential, alternative targets in the therapeutic treatment of androgen independent prostate cancer. Given the fact that many coactivators and corepressors interact with YY1, specific agonists or antagonists to these proteins can be designed and tested, and molecular agents, such as peptide inhibitors, can be used to attenuate AR activity. These studies can potentially lead to the development of new therapeutic strategies that are urgently needed to treat the fatal, advanced prostate cancer.
Abbreviations
| YY1 | = | Yin Yang 1 |
Figures and Tables
Acknowledgements
We thank Ms. Karen Klein (Research Support Core, Wake Forest University Health Sciences) for editing the manuscript. The support for the work came from grants of the Department of Defense (PC060490) and American Cancer Society (RSG-09-082-01-MGO) to Guangchao Sui. The NCI training grant 5T32CA079448-09 supports Paul Cao.
References
- Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 1991; 67:377 - 388
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol 1999; 19:7237 - 7244
- Sui G. The Regulation of YY1 in Tumorigenesis and its Targeting Potential in Cancer Therapy. Mol Cell Pharmacol 2009; 1:157 - 176
- Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function and therapeutic implications in cancer biology. Oncogene 2006; 25:1125 - 1142
- Yant SR, Zhu W, Millinoff D, Slightom JL, Goodman M, Gumucio DL. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res 1995; 23:4353 - 4362
- Deng Z, Wan M, Cao P, Rao A, Cramer SD, Sui G. Yin Yang 1 regulates the transcriptional activity of androgen receptor. Oncogene 2009; 28:3746 - 3757
- Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key?. Gene 1999; 236:197 - 208
- Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci USA 2006;
- Luke MP, Sui G, Liu H, Shi Y. Yin Yang 1 physically interacts with HOXA11 and represses HOXA11-dependent transcription. J Biol Chem 2006;
- Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, et al. Yin Yang 1 is a negative regulator of p53.. Cell 2004; 117:859 - 872
- Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci USA 2004; 101:12165 - 12170
- Santiago FS, Ishii H, Shafi S, Khurana R, Kanellakis P, Bhindi R, et al. Yin Yang-1 inhibits vascular smooth muscle cell growth and intimal thickening by repressing p21WAF1/Cip1 transcription and p21WAF1/Cip1-Cdk4-cyclin D1 assembly. Circ Res 2007; 101:146 - 155
- Palko L, Bass HW, Beyrouthy MJ, Hurt MM. The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J Cell Sci 2004; 117:465 - 476
- Rizkallah R, Hurt MM. Regulation of the transcription factor YY1 in mitosis through phosphorylation of its DNA-binding domain. Mol Biol Cell 2009; 20:4766 - 4776
- Houbaviy HB, Usheva A, Shenk T, Burley SK. Cocrystal structure of YY1 bound to the adenoassociated virus P5 initiator. Proc Natl Acad Sci USA 1996; 93:13577 - 13582
- Lee JS, Galvin KM, See RH, Eckner R, Livingston D, Moran E, et al. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev 1995; 9:1188 - 1198
- Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA 1996; 93:12845 - 12850
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 2004; 18:2627 - 2638
- Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, et al. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev 2003; 17:1019 - 1029
- Deng Z, Wan M, Sui G. PIASy-mediated sumoylation of Yin Yang 1 depends on their interaction but not the RING finger. Mol Cell Biol 2007; 27:3780 - 3792
- Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell 2004; 16:631 - 639
- Chmelar R, Buchanan G, Need EF, Tilley W, Greenberg NM. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. Int J Cancer 2007; 120:719 - 733
- Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer 2004; 4:793 - 805
- Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature 1988; 334:721 - 724
- Castellano G, Torrisi E, Ligresti G, Malaponte G, Militello L, Russo AE, et al. The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle 2009; 8:1367 - 1372
- Zaravinos A, Spandidos DA. Yin yang 1 expression in human tumors. Cell Cycle 2010; 9:512 - 522
- Seligson D, Horvath S, Huerta-Yepez S, Hanna S, Garban H, Roberts A, et al. Expression of transcription factor Yin Yang 1 in prostate cancer. Int J Oncol 2005; 27:131 - 141
- Mousses S, Wagner U, Chen Y, Kim JW, Bubendorf L, Bittner M, et al. Failure of hormone therapy in prostate cancer involves systematic restoration of androgen responsive genes and activation of rapamycin sensitive signaling. Oncogene 2001; 20:6718 - 6723
- Buchanan G, Irvine RA, Coetzee GA, Tilley WD. Contribution of the androgen receptor to prostate cancer predisposition and progression. Cancer Metastasis Rev 2001; 20:207 - 223
- Dehm SM, Tindall DJ. Regulation of androgen receptor signaling in prostate cancer. Expert Rev Anticancer Ther 2005; 5:63 - 74