Abstract
Renin angiotensin system (RAS) is involved in the regulation of cardiovascular homeostasis. Angiotensin (Ang II) is converted from angiotensin I via angiotensin converting enzyme (ACE). Ang II exerts its effects by binding to two types of receptors; AT1R and AT2R. Ang II effect on AT1R promotes proliferation, angiogenesis and metastasis in breast tissues. ACE (I/D) polymorphism is an insertion/deletion of a 287 bp DNA fragment within intron 16 of ACE gene. A1166C is a single nucleotide polymorphism (SNP) in the 3′-UTR of AT1R gene. Both (D) and (C) alleles were found to be related to RAS overactivation.
Subjects and methods
One hundred and twenty postmenopausal Egyptian females were included in the present study and were divided into control group (fifty apparently healthy women) and patients group (seventy breast cancer patients). Detailed history taking was done with stress on age, family history, menstrual, obstetric, medical and drug history. Physical examination including body mass index calculation was done. Histopathological examination was done for tumor grading and staging. Detection of ACE gene (I/D) polymorphism by PCR and AT1R A1166C SNP using PCR/RFLP were done.
Results
A statistically significant difference in AT1R A1166C SNP genotype frequencies was found among the studied groups. The patients group showed higher frequency of “CC” (2.9% vs 0%) and “AC” (44.3% vs 24%) and lower frequency of “AA” genotype (52.9% vs 76%) than controls. The patients also showed significant higher frequency of allele “C” (25% vs 12%) which was associated with increased breast cancer risk with an Odds ratio of 2.4444 (95% CI: 1.1967–4.9931). Testing the dominant model of inheritance revealed a statistically higher frequency of exposed genotypes “AC and CC” among the patients group (47.1% vs 24%, respectively; p = 0.013) with substantial increase in breast cancer risk among the exposed genotypes with an Odds ratio of 2.8243 (95% CI: 1.2679–6.2913). The present study demonstrated that (AC and CC) genotypes of AT1R A1166C SNP and increased BMI can be considered as predictors for breast cancer risk among post menopausal Egyptian females. Results also revealed that A1166C SNP of AT1R gene and ACE/ID polymorphism could not be considered as predictors for breast cancer prognosis.
1 Introduction
The Renin angiotensin system (RAS) is critically involved in the physiological regulation of blood pressure, volume homeostasis and tissue perfusion. It plays an integral role in the pathogenesis of hypertension and other cardiovascular diseases.Citation1,Citation2 RAS is now well recognized as a dual vasoactive system, acting as both a circulating endocrine system and a local tissue system.Citation3,Citation4
Angiotensin converting enzyme (ACE or Peptidyl-Dipeptidase A) is a zinc dependant dipeptidyl carboxypeptidase that cleaves a dipeptide from the carboxyl terminus of the decapeptide Ang I to form Ang II. In addition ACE metabolizes a number of other peptides, including the vasodilator peptide bradykinin, to inactive metabolites. Thus, functionally ACE has a dual enzymatic action resulting in increased vasoconstriction and decreased vasodilation.Citation5
Ang II is a potent vasoconstrictor which acts by binding to three recognized angiotensin receptor subtypes, angiotensin receptor type 1 (AT1R) and the angiotensin receptor type 2 (AT2R), both are structurally similar, and angiotensin receptor type 4 (AT4R) which is different.Citation6,Citation7 Binding of Ang II to AT1R stimulates vasoconstriction, release of aldosterone, angioneogensis, cell growth and proliferation while its binding to AT2R causes growth inhibition, apoptosis and vasodilatation.Citation7,Citation8
In human, the gene encoding ACE is located on the long arm of chromosome 17 (17q23). The gene is 21 kilo bases (kb) long and comprises 26 exons and 25 introns. The most popular ACE gene polymorphism is an insertion/deletion (I/D) of a 287-base-pair DNA fragment within the intron 16 of the ACE gene (NCBI Ref. SNP ID: http://wb-strain:rs1799752). This polymorphism was found to be linked to the ACE activity level which is doubled in homozygous deletion carriers (DD) when compared to II carries, while ID carriers show intermediate activity.Citation9–Citation10Citation11
The human AT1R gene is mapped to chromosome 3q21–q25 and spans more than 55 kb of genomic DNA. A1166C (NCBI Ref. SNP ID: http://wb-strain:rs5186) is a single nucleotide polymorphism (SNP) in which there is an A/C transversion at position 1166 in the 3′ untranslated region (3′-UTR) of AT1R gene. It has been found that the A1166C SNP occurs in a cis-regulatory site in the 3′-UTR of AT1R gene which after transcription is recognized by a specific microRNA (miR-155).Citation9
Micro RNAs (miRNAs) are a class of small endogenous non coding single stranded RNAs (19–24 nucleotides) that are involved in post transcriptional gene regulation. Micro RNAs regulate gene expression through binding to their target messenger RNAs (mRNAs) in the 3′-UTR, by a partial base-pairing mechanism. This binding would result in either inhibition of translation or induction of target mRNA degradation.Citation11,Citation12 In AT1R gene; the A1166C SNP is considered one of the miRSNPs (SNP in a micro RNA target site) that alter a DNA sequence which when transcribed will form a part of a cis-regulatory site located in the AT1R mRNA 3′-UTR, at a site where miR-155 is known to interact. When A allele is present in this regulatory site, there will be at least seven consecutive base pair regions of complementarity between the 5′ end of miR-155 and the AT1R mRNA target site, thus enabling miR-155 to interact with this regulatory site thus inhibiting AT1R mRNA translation. On the other hand, when the C-allele is present, base-pairing complementarity is interrupted, and the ability of miR-155 to interact with the cis-regulatory site is decreased. As a consequence, miR-155 can no longer attenuate translation, resulting in increased AT1R densities leading to an increase in RAS activation.Citation9,Citation11,Citation13
The RAS, in particular the AT1R, is often up-regulated during the progression from normal to malignant phenotypes, indicating a possible correlation between the RAS and tumor progression.Citation14,Citation15 The RAS plays a role in modulation of angiogenesis, cellular proliferation, immune responses, inflammation and extracellular matrix formation. Manipulation of the RAS may, therefore, provide a safe and inexpensive anticancer strategy.Citation16–Citation17Citation18
In Egypt, breast cancer represents 24% of total cancer cases (37% in women and 0.8% in men) according to latest records of Egypt National Cancer Institute (NCI) series of 55,740 patients between 2002 and 2007.Citation17 The three strongest prognostic determinants in operable breast cancer are lymph node (LN) stage, primary tumor size, and tumor histologic grade, they were combined in an index termed Nottingham prognostic index (NPI).Citation18
It has been suggested that a positive correlation might exist between polymorphisms of genes that code for proteins of the RAS and the risk of developing breast cancer.Citation19,Citation9 So far, there are no data available in the literature regarding the association of AT1R receptor (A1166C) and ACE gene (I/D) polymorphisms in Egyptian women with breast cancer, the aim of the study is assessment of these polymorphisms in Egyptian females with breast cancer.
Aim of the work: Studying any possible association between the polymorphisms of angiotensin converting enzyme (I/D) and angiotensin II type I receptor (A1166C) and breast cancer among post menopausal Egyptian females.
Subjects and methods: After the acceptance of the Ethics Committee of the Medical Research Institute (MRI), one hundred and twenty Egyptian females were recruited from the outpatient clinic of the MRI hospital, they were divided as follows: Group I (control group): It included fifty apparently healthy postmenopausal Egyptian females without any family history of breast cancer. Group II (patients group): it included seventy postmenopausal Egyptian females with breast cancer with positive family history. Exclusion criteria: patients taking ACE inhibitors (ACEIs), angiotensin II receptor blockers (ARBs) or any drugs that affect ACE or angiotensin II level, patients with history or clinical evidence of any other malignancy and patients with liver, renal and lung diseases.
An informed consent was taken from all subjects included in this study before its start.
All studied subjects were subjected to the following: Detailed history taking with special stress on age, age of onset of breast cancer, family history, menstrual and obstetric history including age of menarche, age of menopause, parity, medical history, drug history especially intake of antihypertensive drugs, oral contraceptive pills (OCPs) and hormonal replacement therapy (HRT). Thorough physical examination including estimation of body mass index (BMI). Histopathological examination of the breast tissues was done for tumor grading and histological staging.Citation20,Citation21
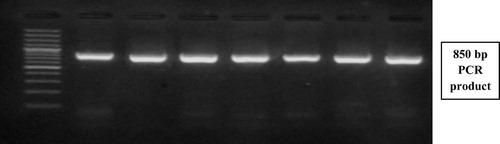
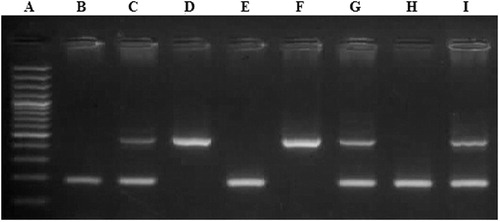
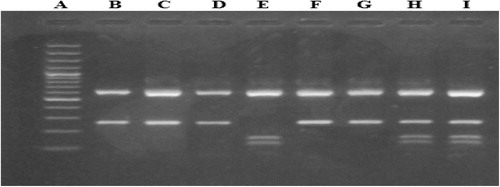
Laboratory investigations: After 10 h fasting; five milliliters of venous whole blood was withdrawn from each subject and the following parameters were performed: serum glucose, urea and creatinine concentrations, serum activity of alkaline phophatase, alanine amino transferase and aspartate amino transferase were conducted on Olympus AU400 clinical chemistry analyzer (Beckman Coulter Inc.).Citation22 Serum CA 15.3 was done on IMMULITE 1000, using a two-step sequential chemiluminescent immunometric assay Citation23 and molecular studies were done which included: DNA extraction from peripheral blood leucocytes, followed by detection of the presence of the (I) and (D) alleles in the ACE gene by Polymerase chain reaction (PCR) amplification using specific primers (Forward primer: 5′-CTG GAG ACC ACT CCC ATC CTT TCT-3′, Reverse primer: 5′-GAT GTG GCC ATC ACA TTC GTC AGA T-3′) followed by agarose gel electrophoresis for the PCR product see (). All samples found to be (DD) after amplification with the conventional primers, were reamplified using allele specific primer pair which recognized insertion specific sequences (Forward primer: 5′-TGG GAC CAC AGC GCC CGC CAC TAC-3′, Reverse primer: 5′-TCG CCA GCC CTC CCA TGC CCA TAA-3′). Allele (Insertion) Specific PCR was done to avoid mistyping of the (I) allele as (D) allele in (ID) heterozygous carriers see (). Detection of AT1R (A1166C) gene polymorphism using polymerase chain reaction/restriction fragment length polymorphism (PCR/RFLP) technique was done as follows: PCR amplification using specific primers (Forward primer: 5′-AAT GCT TGT AGC CAA AGT CAC CT-3′, Reverse primer: 5′-GGC TTT GCT TTG TCT TGT TG-3′) see (), restriction digestion of PCR products using Dde-I enzyme and agarose gel electrophoresis of digested PCR products to detect A1166C gene polymorphism see ().

2 Statistical analysisCitation24
Statistical analysis was done using SPSS program version 20 (Statistical Package of social sciences, Chicago, USA)Citation25. For quantitative variables: D’Agostino – Pearson K-squared test for normality was used to test for the degree of deviation from normal distribution across all quantitative variables in all groups and subgroups. For qualitative variables: The data of the qualitative variables were summarized in the form of frequency or percentages. The Chi-Square test (χ2 test) with a Monte Carlo estimate of the exact p-value, was used to compare proportions of nominal clinical data variables between groups for 2 by 3 table or more (not for 2 × 2 tables). Chi-square test was performed only if at least 80% of the cells have an expected frequency of 5 or greater, and no cell has an expected frequency smaller than 1. For 2 × 2 tables, Fisher’s Exact Test was used to compare proportions of nominal clinical data variables. Fisher’s Exact Test can only be applied to 2 by 2 tables whatever the cell values are. For 2 or more by 3 or more tables with less than 80% of the cells having an expected frequency of ⩾5, or one or more cells having an expected frequency <1, neither the Chi-square test nor the normal Fisher’s exact test could be used. In this situation the Freeman–HaltonCitation26 extension of the Fisher’s exact probability test (Fisher–Freeman–Halton’s test) was used instead. p-Values of less than 0.05 was considered a statistically significant difference.
Testing deviation from the HWE is generally performed using Pearson’s Chi-Squared goodness of fit test, using the observed genotype frequencies obtained from the data and the expected genotype frequencies obtained using the HW equations.Citation27
Odds ratio and risk assessment:Citation28 To measure the effect or the impact of A1166C AT1R SNP and ACE I/D genotypes or alleles on the risk of developing breast cancer.
Logistic regression analysis: Used to determine the relationship between predictor variables and a dichotomously coded dependent variable.Citation29
3 Results
A statistically significant difference (p = 0.0211∗) in the AT1R A1166C SNP genotype frequencies between breast cancer patients and control groups was found. The breast cancer patients had a higher frequency of the homo-mutant genotype “CC” than controls (2.9% vs 0%), a higher frequency of the hetero-mutant genotype “AC” than controls (44.3% vs 24%) and a lower frequency of the homo-wild genotype “AA” than controls (52.9% vs 76%) ().
Table 1 Comparison of AT1R A1166C SNP genotype frequencies among the studied groups.
There was a statistically significant (p = 0.0134∗) difference in the A1166C AT1R SNP allele frequencies between breast cancer patients and controls, while the patients had statistically significant higher frequency of the allele “C” than controls (25% vs 12%) and allele C was associated with a statistically increased breast cancer risk with an Odds ratio of 2.4444 (95% CI: 1.1967–4.9931) ().
Table 2 Comparison of AT1R A1166C SNP allele frequencies among the studied groups.
Testing the recessive model was not applicable due to the low number of CC genotypes; also the Odds ratio cannot be estimated because of the absence of CC genotype among the control subjects.
Testing the dominant model of inheritance revealed a statistically higher frequency of exposed genotypes “AC and CC” among the breast cancer patients group when compared to control group (47.1% vs 24%, respectively; p = 0.013) with substantial increase in breast cancer risk among the exposed group “AC and CC” with an Odds ratio of 2.8243 (95% CI: 1.2679–6.2913) when compared to unexposed group “AA” (). Only BMI and A1166C AT1R SNP genotype were left as significant predictors in this logistic regression model. The Wald criterion demonstrated that both BMI and A1166C AT1R SNP genotype made a significant contribution to the prediction of breast cancer risk (p-value = 0.001 and 0.009, respectively) ().
Table 3 Assessment of the risk of developing breast cancer according to AT1R + A1166C SNP genotypes in different models of inheritance.
Table 4 Variables included in the final logistic regression model with breast cancer risk as the dependent variable.
No statistically significant difference was found (p = 0.940) in the ACE I/D genotype frequencies between the studied groups (). No statistically significant difference (Exact p-value = 0.893, Odd’s ratio = 0.9353 with 95% confidence interval: 0.5507–1.5886) in the ACE I/D allele frequencies was found among the studied groups ().
Table 5 Comparison of ACE I/D genotype frequencies among the studied groups.
Table 6 Comparison of ACE I/D allele frequencies among the studied groups.
4 Discussion
Breast cancer accounts for 18% of all female cancers worldwide and it is one of the main causes of global health burden. Several environmental, anthropometric, and genetic factors could contribute to increased risk of breast cancer. A number of genetic variants have been identified to be potentially associated with breast cancer risk.Citation30
There is now increasing evidence that RAS may have local tissue actions involving angiogenesis, cellular proliferation, apoptosis and inflammation. A polymorphic variant composed of an insertion (I) or a deletion (D) of a 287 base pair insert in intron 16 (rs: 1799752) has been identified to be associated with altered concentration of ACE.Citation31 The DD genotype is associated with the highest serum levels of the enzyme, the ID genotype with the intermediate levels and the II genotype with the lowest levels.Citation32,Citation33
A1166C is a single nucleotide polymorphism (SNP) in which there is an A to C transversion at position 1166 in the 3′ untranslated region of the AT1R gene. When the C-allele is transcribed the ability of micro-RNA 155 (miR-155) to attenuate AT1R mRNA translation is interrupted, resulting in an overall increase in AT1R density and thus increasing the biological actions of Ang II resulting in RAS over activation.Citation34,Citation37
In the present study, the association between ACE I/D and AT1R A1166C SNP and breast cancer was studied in a sample of one hundred and twenty Egyptian females divided into two groups; The control group (Group I) included fifty apparently healthy postmenopausal Egyptian females without family history of breast cancer; their mean age was 62.74 ± 5.47 years. The patient group (Group II) included seventy postmenopausal Egyptian females with breast cancer; their mean age was 52.1 ± 5.79 years.
Comparing the genotype frequencies of ACE (I/D) polymorphism among the studied groups revealed no statistically significant difference (; p = 0.940) between the control group (II = 16%, ID = 42%, DD = 42%) and breast cancer patients group (II = 18.6%, ID = 40%, DD = 41.4%). Furthermore, there was also no statistically significant difference in the allelic frequencies of ACE (I/D) polymorphism between control group (“I” allele = 37%, “D” allele = 63%) and breast cancer patients group (“I” allele = 39%, “D” allele = 61%) (; p = 0.893, OR = 0.9353; 95% CI: 0.5507–1.5886). The genotype distribution as well as allele frequencies of ACE (I/D) polymorphism were both found to be in agreement with Hardy–Weinberg equilibrium. Several studies tried to explore the association between ACE ID polymorphism and breast cancer risk; however they revealed conflicting results. Some studiesCitation30,Citation35 reported that D allele was positively associated with breast cancer risk; another studyCitation36 reported that D allele was negatively associated with breast cancer risk; and others Citation37 (in agreement with the present study) found no significant difference suggesting that ACE I/D polymorphism may not be a good predictor for breast cancer risk.
Koh et al.Citation35 first studied both I/D and A240T polymorphisms of the ACE gene in association with breast cancer, they found that women with both A and I alleles had a statistically significant reduction of breast cancer risk compared with those carrying either the TT and/or DD genotypes (the high-activity genotype) (OR: 0.46; 95% CI: 0.27–0.81). González-Zuloeta Ladd et al.Citation30 conducted a prospective cohort study to evaluate relationship of the ACE I/D polymorphism with breast cancer risk in 4117 Caucasian postmenopausal women. They found that DD carriers showed a significantly increased risk of developing breast cancer when compared with the II carriers (OR: 1.86; 95% CI: 1.06–3.27). This association remained after performing logistic regression analysis to adjust for other risk factors, including body mass index, age at menarche, age at menopause, hormone replacement therapy, and hypertension. In agreement with the previous studies; Van der Knaap et al.Citation38 found that carriers of the high activity genotype DD had an increased risk of breast cancer compared with low-activity II/ID genotype carriers (hazard ratio [HR]: 1.47; 95% CI: 1.05–2.04), but no association was demonstrated for other cancers.
On the other hand Haiman et al.Citation36 studied ACE I/D polymorphisms in relation to breast cancer risk among different ethnic groups. They found that D allele was reversely associated with breast cancer risk. (OR: 1.30; 95% CI: 1.05–1.61), although associations were not entirely consistent across ethnic groups suggesting that D allele may be a marker of low ACE levels in some populations, but not all populations.
In agreement with the present study; Sun et al.Citation37 conducted a recent meta analysis which included four studies (cases: 1422; controls: 3044), they studied the association between ACE (I/D) polymorphism and breast cancer. The results of this meta analysis showed that there was no significant difference in genotype distribution (DD, ID or II) between breast cancer patients and controls. They suggested that ACE I/D polymorphism may not be a genetic risk factor for breast cancer.
These discrepancies in results might be due to insufficient statistical power and lack of adjustment for other potential breast cancer confounding factors in some studies, recruitment procedures of the study population, genetic heterogeneity and differences in linkage disequilibrium between different populations as well as environmental backgrounds. Another reason could be the differences in population characteristics (e.g., use of ACE inhibitors and green tea intakeCitation39 could influence the association between RAS genes and breast carcinogenesis), these differences in patient characteristics are likely to influence their phenotypes and may have an impact on the study results. Using different methods and techniques with different performance characteristics (e.g., insertion specific amplification to avoid ID mistyping as DD was not done in all studies) could be another possible reason for result inconsistency.
As for A1166C SNP of AT1R gene, comparing the genotype frequencies among the studied groups () revealed a statistically significant difference (p-value = 0.0211∗) between control group and breast cancer patients group, where the breast cancer patients have a higher frequency of the homozygous mutant genotype (CC) than controls (2.9% vs 0%), a higher frequency of the heterozygous mutant genotype (AC) than controls (44.3% vs 24%) and a lower frequency of the homozygous wild genotype (AA) than controls (52.9% vs 76%). Furthermore, there was also a statistically significant difference (p = 0.0134) in the allelic frequencies of A1166C SNP of AT1R gene between control group and breast cancer patients group, where the breast cancer patients had a higher frequency of the mutant allele (C) than controls, and a lower frequency of the wild allele (A) than controls (OR 2.4444; 95% CI: 1.0616–4.0233) (). From these results it could be concluded that the mutant allele (C) was associated with a statistically significant increased risk of developing breast cancer. The genotype distribution as well as allele frequencies of A1166C SNP of AT1R gene were both found to be in agreement with Hardy–Weinberg equilibrium.
Assuming a recessive model of inheritance for AT1R A1166C SNP; the exposed group is only subjects with the (CC) genotype while the unexposed group is subjects with both (AC and AA) genotypes combined. Unfortunately, in the present study the Odds ratio involving the (CC) genotype alone could not be estimated because of the absence of this genotype among the control subjects. Accordingly assessing the risk in the recessive model was not possible. Results of the present study revealed also a statistically higher frequency of exposed genotypes “AC and CC” among the breast cancer patients group when compared to control group (p = 0.013) with substantial increase in breast cancer risk among the exposed group (AC and CC) with an OR of 2.8243 (95% CI: 1.2679–6.2913) when compared to unexposed group (AA) (). This finding remains after adjustment for other breast cancer confounding factors, the variables included in the final logistic regression model with breast cancer risk as the dependent variable were BMI (p-value: 0.001, Exp (B): 1.265, 95% CI: 1.102–1.452) and A1166C AT1R SNP genotype (AC and CC vs AA) (p-value: 0.009, Exp (B): 3.419, 95%CI: 1.364–8.569) ().
The findings of the present study could be explained by the ability of AT1R A1166C SNP to cause RAS overactivation. Since A1166C polymorphism occurs in the 3′-UTR of the human AT1R gene, the biological mechanism by which this SNP could possibly lead to RAS overactivation has been always questionable. Martin et al.Citation11 and Sethupathy et al.Citation12 demonstrated that A1166C SNP is one of the miRSNPs that alter a DNA sequence which when transcribed will form a part of a cis-regulatory site located in the AT1R mRNA 3′-UTR, at a site where miRNA “miR-155” is known to interact. When the (C) allele is present, base-pairing complementarity is interrupted, and the ability of miR-155 to interact with this cis-regulatory site is decreased. As a consequence, miR-155 can no longer attenuate translation, resulting in increased AT1R densities leading to an increase in RAS activation.Citation40 Increased AT1R mRNA transcription was observed in breast cancer cells when compared to normal cells. This will subsequently lead to enhanced tumurogenic actions of Ang II on breast cells including angiogenesis, cell proliferation, and inflammation. Since AT1R A1166C SNP is known to increase AT1R mRNA expression, therefore this SNP might be related to increased AT1R expression in breast cancer cells.Citation41,Citation42
So far, only limited studies have evaluated the association between AT1R A1166C SNP and breast cancer risk, yet they also yielded inconsistent results. Alves Corrêa et al.Citation43 and Namazi et al.Citation33 suggested that the A1166C polymorphism was not associated with breast cancer risk (p-values: 0.114 and 0.86, respectively). In contrast, Mendizábal-Ruiz et al.Citation44 found that the frequency of (A) allele was higher in breast cancer patients group when compared to controls (p = 0.036) and that the (C) allele carriers had reduced risk of breast cancer (OR: 0.53, p-value = 0.0356). Recently; Xi et al.Citation45 evaluated the three published studies (235 cases and 601 controls). Significant association between AT1R A1166C SNP and breast cancer risk was observed for (AC) versus (AA) and dominant model. However, after excluding one study that was not in Hardy–Weinberg equilibrium (by Alves Corrêa et al.Citation43, this association disappeared while a marginally significant association was observed for CC versus AA (OR = 0.31, 95% CI 0.10–0.99). Since publication bias was not assessed for these findings; they recommended further studies to explore the association between this SNP and breast cancer risk.
From the previously mentioned studies, only limited data are available regarding the association between AT1R A1166C SNP and breast cancer risk. Lack of consistency in the findings, limited sample size, insufficient statistical power for one study as well as paucity of explanations for the observed associations could be noticed. Further large population based studies are still needed to get a conclusive evidence about the association between AT1R A1166C SNP and the risk of breast cancer, as well as AT1R expression in breast cancer tissues in relation to this SNP genotype.
In conclusion, the present study demonstrated that (AC and CC) genotypes of AT1R A1166C SNP together with increased BMI can be considered as significant predictors for breast cancer risk among post menopausal Egyptian Females. Results also revealed that A1166C SNP of AT1R gene and ACE/ID polymorphism might not be a good predictor for breast cancer prognosis. Also, pharmacological inhibition of the angiotensin II carcinogenic effect through inhibition of AT1R together with weight reduction could be used to delay or prevent the occurrence of breast cancer in post menopausal females, although more detailed trials and experiments are required to confirm this conclusion.
Conflict of interest
None.
Notes
Peer review under responsibility of Alexandria University Faculty of Medicine.
Available online 7 January 2014
References
- A.Nguyen Dinh CatR.M.TouyzA new look at the renin-angiotensin system-focusing on the vascular systemPeptides32201121412150
- ARB Trialists CollaborationEffects of telmisartan, irbesartan, valsartan, candesartan, and losartan on cancers in 15 trials enrolling 138, 769 individualsJ Hypertens292011623635
- M.BaderD.GantenUpdate on tissue renin-angiotensin systemsJ Mol Med862008615621
- J.L.ZhuoX.C.LiNew insights and perspectives on intrarenal reninangiotensin system: focus on intracrine/intracellular angiotensin IIPeptides32201115511565
- R.M.CareyH.M.SiragyNewly recognized components of the renin-angiotensin system: potential roles in cardiovascular and renal regulationEndocr Rev242003261271
- J.W.WrightB.J.YamamotoJ.W.HardingAngiotensin receptor subtype mediated physiologies and behaviors: new discoveries and clinical targetsProg Neurobiol842008157181
- A.MuscellaS.GrecoM.G.EliaC.StorelliS.B.MarsiglianteAngiotensin II stimulation of Na+/K+ATPase activity and cell growth by calcium-independent pathway in MCF-7 breast cancer cellsJ Endocrinol1732002315323
- S.NouetC.NahmiasSignal transduction from the angiotensin II AT2 receptorTrends Endocrinol Metab11200016
- X.JeunemaitreGenetics of the human renin angiotensin systemJ Mol Med862008637641
- F.A.Sayed TabatabaeiB.A.OostraA.IsaacsC.M.van DuijnJ.C.WittemanACE polymorphismsCirc Res98200611231133
- M.M.MartinJ.A.BuckenbergerJ.JiangG.E.MalanaG.J.NuovoM.ChotaniThe human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates micro RNA-155 bindingJ Biol Chem2823320072426224269
- P.SethupathyC.BorelM.GagnebinG.R.GrantS.DeutschT.S.EltonHuman microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypesAm J Hum Genet8122007405413
- J.BrenneckeA.StarkR.B.RussellS.M.CohenPrinciples of microRNA-target recognitionPLoS Biol32005e85
- G.R.SmithS.MissailidisCancer, inflammation and the AT1 and AT2 receptorsJ Inflamm1120043
- M.TahmasebiS.BarkerJ.R.PuddefootG.P.VinsonLocalisation of renin-angiotensin system (RAS) components in breastBr J Cancer95120066774
- M.FujitaI.HayashiS.YamashinaM.ItomanM.MajimaBlockade of angiotensin AT1a receptor signaling reduces tumor growth, angiogenesis, and metastasisBiochem Biophys Res Commun29422002441447
- Ali Eldin N. Cancer Statistics 2002–2007, National Cancer Institute, Egypt 2010. Department of Biostatistics and Cancer Epidemiology. Preliminary report.
- E.A.RakhaM.E.El-SayedA.H.LeeC.W.ElstonM.J.GraingeZ.HodiPrognostic significance of Nottingham histologic grade in invasive breast carcinomaJ Clin Oncol2619200831533158
- J.K.LohGene polymorphisms in the renin-angiotensin-aldosterone system and breast carcinogenesis: is there a connection?TSMJ920084851
- E.A.RakhaJ.S.Reis-FilhoF.BaehnerD.J.DabbsT.DeckerV.EusebiBreast cancer prognostic classification in the molecular era: the role of histological gradeBreast Cancer Res1242010207
- National Comprehensive Cancer Network (NCCN) guidelines. Breast Cancer Version 2.2011, pp. ST1-4.
- C.A.BurtisE.R.AshwoodD.E.BrunsTietz text book of clinical chemistry and molecular diagnostics5th ed.2012Elsevier Saunders Company. St Louis [pp.718-21, 685–6, 680–4, 578–80 and 575–6, respectively]
- P.R.SlevM.L.RawlinsW.L.RobertsPerformance characteristics of seven automated CA 15-3 assaysAm J Clin Pathol12552006752757
- L.E.DalyG.J.BourkeInterpretation and uses of medical statistics5th ed.2000Oxford, Blackwell ScienceMalden, MA
- B.K.PuriSPSS in practice: an illustrated guide2nd ed.2002ArnoldLondon; New York
- S.LydersenV.PradhanP.SenchaudhuriP.LaakeChoice of test for association in small sample unordered r × c tablesStat Med26200743284343
- S.RodriguezT.R.GauntI.N.DayHardy–Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studiesAm J Epidemiol16942009505514
- B.PietaD.SamulakT.OpalaM.WilczakS.Grodecka-GazdeckaK.Wieznowska-MaczyńskaAnalysis of odds ratio of increased relative risk of developing breast cancer in different groups of womenEur J Gynaecol Oncol31120105054
- C.J.PengK.L.LeeG.M.IngersollAn introduction to logistic regression analysis and reportingJ Educ Res9612002314
- A.M.González-Zuloeta LaddA.Arias VásquezF.A.Sayed-TabatabaeiJ.W.CoeberghA.HofmanO.NjajouAngiotensin-converting enzyme gene insertion/deletion polymorphism and breast cancer riskCancer Epidemiol Biomarkers Prev149200521432146
- H.JiaB.WangL.YuZ.JiangAssociation of angiotensin-converting enzyme gene insertion/deletion polymorphism with polycystic ovary syndrome: a meta-analysisJ Renin Angiotensin Aldosterone Syst1432013255262
- S.YigitA.InanirS.TuralO.AtesAssociation of angiotensin converting enzyme (ACE) gene I/D polymorphism and rheumatoid arthritisGene51112012106108
- S.NamaziA.MonabatiS.Ardeshir-Rouhani-FardN.AzarpiraAssociation of angiotensin I converting enzyme (insertion/deletion) and angiotensin II type 1 receptor (A1166C) polymorphisms with breast cancer prognostic factors in Iranian populationMol Carcinog4912201010221030
- K.ZhangB.ZhouL.ZhangAssociation study of angiotensin II type 1 receptor: A1166C (rs5186) polymorphism with coronary heart disease using systematic meta-analysisJ Renin Angiotensin Aldosterone Syst1422013181188
- W.P.KohJ.M.YuanC.L.SunD.van den BergA.SeowH.P.LeeAngiotensin I-converting enzyme (ACE) gene polymorphism and breast cancer risk among Chinese women in SingaporeCancer Res6332003573578
- C.A.HaimanS.O.HendersonP.BretskyL.N.KolonelB.E.HendersonGenetic variation in angiotensin I-converting enzyme (ACE) and breast cancer risk: the multiethnic cohortCancer Res6320200369846987
- M.SunC.LiuF.WeiJ.ZhongY.SunAssociation of angiotensin I converting enzyme insertion/deletion polymorphism with breast cancer: a meta-analysisJ Renin Angiotensin Aldosterone Syst1242011611616
- R.Van der KnaapC.SiemesJ.W.CoeberghC.M.van DuijnA.HofmanB.H.StrickerRenin-angiotensin system inhibitors, angiotensin I-converting enzyme gene insertion/deletion polymorphism, and cancer: the Rotterdam studyCancer11242008748757
- J.M.YuanW.P.KohC.L.SunH.P.LeeM.C.YuGreen tea intake, ACE gene polymorphism and breast cancer risk among Chinese women in SingaporeCarcinogenesis268200513891394
- H.S.LimJ.Y.ChoD.S.OhJ.Y.ChungK.S.HongK.S.BaeAngiotensin II type 1 receptor 1166A/C polymorphism in association with blood pressure response to exogenous angiotensin IIEur J Clin Pharmacol6320071726
- A.JethonB.PulaA.PiotrowskaA.WojnarJ.RysP.DziegielAngiotensin II type 1 receptor (AT-1R) expression correlates with VEGF-A and VEGF-D expression in invasive ductal breast cancerPathol Oncol Res1842012867873
- S.Rodrigues-FerreiraM.AbdelkarimP.Dillenburg-PillaA.C.LuissintA.di-TommasoF.DeshayesAngiotensin II facilitates breast cancer cell migration and metastasisPLoS ONE742012e35667
- S.A.Alves CorrêaS.M.Ribeiro de NoronhaN.C.Nogueira-de-SouzaC.Valleta de CarvalhoA.M.Massad CostaJ.Juvenal LinharesAssociation between the angiotensin-converting enzyme (insertion/deletion) and angiotensin II type 1 receptor (A1166C) polymorphisms and breast cancer among Brazilian womenJ Renin Angiotensin Aldosterone Syst10120095158
- A.P.Mendizábal-RuizJ.MoralesX.Castro MartinezS.A.Gutierrez RubioL.ValdezJ.G.Vásquez-CamachoRAS polymorphisms in cancerous and benign breast tissueJ Renin Angiotensin Aldosterone Syst12220118592
- B.XiT.ZengL.LiuY.LiangW.LiuY.HuAssociation between polymorphisms of the renin-angiotensin system genes and breast cancer risk: a meta-analysisBreast Cancer Res Treat13022011561568