Highlights
| • | SYBR Green offers good level of sensitivity and specificity. | ||||
| • | SYBR Green assay is suitable for large-scale screening of donated blood. | ||||
| • | SYBR Green PCR has the advantage of low cost. | ||||
Abstract
The prevalence of hepatitis C virus (HCV) is highest in Egypt compared to other countries. Nucleic acid amplification test (NAT) allows detection of HCV early during the course of infection. Unfortunately, NAT is more expensive than ELISA, thus its routine use as a screening tool for blood products or in clinical practice is quite limited. The aim of this study was to compare two common RT-PCR methods, TaqMan probe technique and SYBR Green method in quantitative detection of HCV RNA for diagnosis and follow up of HCV patients. Among the recruited 220 HCV patients, 154 (70%) were HCV-RNA positive by both the techniques, while 24 (10.9%) were negative by both techniques. On the other hand, 40 (18.2%) cases were HCV RNA positive only by SYBR Green technique, and 2 (0.9%) only by TaqMan probe technique. Forty (20.4%) of the 196 chronic HCV cases were HCV-RNA positive by SYBR Green but negative by TaqMan probe technique.
Conclusion
This method is useful for rapid qualitative detection of HCV infection and particularly suitable for routine diagnostic applications.
Keywords:
1 Introduction
Hepatitis C virus (HCV) causes both acute and chronic infection. Acute HCV infection is usually asymptomatic, and very rarely associated with a life-threatening condition. About 15–45% of the infected individuals naturally clear the virus within six months of infection without any medical intervention, and in only 55–85% of individuals the infection takes into chronic form. In chronic HCV infection, the risk of liver cirrhosis is 15–30% within 20 years. At a global level, 130–150 million people are having chronic hepatitis C infection.Citation1 Since most countries do not have complete data, this estimate is based on weighted average for regions rather than individual countries. Its lowest prevalence is in the United Kingdom and Scandinavia, while the highest has been reported (15–20%) from Egypt.Citation2
The tests that can detect antibody to HCV (anti-HCV) were first licensed by the Food and Drug Administration (FDA) in 1990. Since then, anti-HCV tests are being widely used for clinical diagnosis and screening of asymptomatic persons.Citation3
CDC has recommended that a person should be considered to have serologic evidence of HCV infection only after the positive result of an anti-HCV screening-test has been verified by a more specific serologic test (e.g., recombinant immunoblot assay (RIBA) or nucleic acid test (NAT).Citation4
The assays for antiviral antibodies, cannot detect an early HCV infection; this leads to false negative result. Nucleic acid amplification tests (NAT) allow detection of HCV much earlier in the course of infection, often as early as 11 days. These tests can also differentiate between active and resolved infection. In addition, NAT has a very high sensitivity and specificity, thus it can be used as an independent method for confirming HCV infection.Citation5–Citation7 Unfortunately NAT is more expensive than ELISA, thus its routine use as a screening tool for blood products or in clinical practice is limited. Several methods, such as branched DNA (signal amplification), transcription-mediated amplification and PCR, are currently in use for detection of HCV RNA.Citation8
Conventional PCR uses gel electrophoresis to identify the amplification product that takes several hours. A cross-contamination of HCV nucleic acid between the specimens may occur. Recently, PCR has been combined with fluorescence dye technologies that offer real-time detection of products during the amplification. This combination allows convenient detection of PCR products and obviates the need for extensive assessment procedures.Citation9
HCV RT-PCR per sample costs in different private laboratories in Egypt range from $25 to $40, while SYBR Green I assay costs $15 to obtain the same results.
SYBR Green I when binds to the minor groove of double-stranded deoxyribonucleic acid (dsDNA) emits fluorescence 1000-fold greater than it's free in solution. Therefore, the increasing amount of dsDNA present in the reaction tube lead to the greater amount of bound dyes and increasing fluorescent signal from SYBR Green I. This method is relatively cost benefit and easy to use. Specificity is the most important concern with the usage of any of these non-specific dsDNA-binding dyes. Non-specific products reflected in dissociation curve of the amplified product as non-specific peaks.Citation7
The aim of this study was to compare two common RT-PCR methods, TaqMan probe technique and SYBR Green method in quantitative detection of HCV RNA for diagnosis and follow up of HCV patients.
2 Materials and methods
Two hundred and twenty consecutive HCV patients, naïve or under treatment, were selected from the outpatient clinics of the Medical Research Institute, Alexandria University. Exclusion criteria included patients with renal, cardiac, neoplastic disease, immunological disorders. Each patient responded to a questionnaire to collect demographic data such as age and sex.
The study protocol was approved by the Institutional Review Board and Ethics Committee of the Medical Research Institute, Alexandria University. All patients signed an informed consent form.
2.1 RNA extraction
RNA extraction was carried out from 140 µL of serum specimens using a QIAamp viral RNA kit (QIAGEN, Valencia, CA) following the manufacturer’s instructions. The extract was divided into two tubes; one for the TaqMan technique and the other stored at −20 °C to be used for SYBR Green technique (which was performed immediately after the TaqMan PCR within 3 hoursto avoid RNA degradation).
2.2 Determination of viral load using TaqMan technique
The extracted RNA (10 μL) was amplified by using HCV Qiagen kit (Artus, QS-RGQ(72) ref. 4518366 HCV PCR kit). RT-PCR reactions were performed in a final volume of 25 μL using the PCR MX3000 Stratagene system (Applied Biosystems Inc., Foster city, California, USA).
For a PCR reaction the following thermal profile was employed: 50 °C for 30 min (reverse transcriptase step) followed by AmpliTaq activation step at 95 °C for 15 min, followed by a three-step PCR protocol, 95 °C for 30 s and followed by annealing at 50 °C for 1 min and extension at 72 °C for 30 s for 40 cycles with end point fluorescence detection.
For determination of the viral load, Ct value was used against standard curve to determine the viral load. Four standard HCV copy DNA molecules (provided by the manufacturer) were processed as described and the standard curve was plotted.
2.3 Detection of HCV RNA by SYBR Green technique
SYBR Green I real-time RT-PCR was carried out using Sigma-Fermentas one step RT PCR kit in a 25 μL reaction volume that contained; 12.5 µL SYBR Green master mix, 1.25 µL RT enhancer, 0.3 µL Forward primer (k 78), 0.3 µL Reverse primer (k 80), 0.25 µL RT (reverse transcriptase), 10 µL extract, and 0.65 µL PCR grade water.
Real-time PCR was performed by using the Mx3000P Stratagene real-time PCR system. The RT-PCR protocol included an RT (reverse transcription) step for 30 min at 50 °C and enzyme activation step for 15 min at 95 °C, followed by 40 cycles of denaturation (95 °C for 15 s), annealing (55 °C for 30 s), and extension (72 °C for 30 s).
Single fluorescence detection was performed in each cycle at 72 °C to reveal the positive samples. Melting curve analysis was done immediately after the final amplification step by heating the samples at 95 °C for 60 s, cooling to 55 °C for 30 s, and heating to 95 °C for 30 s with continuous fluorescence recording. Melting curves were recorded by plotting fluorescence signal intensity versus temperature. The complete procedure included amplification, data acquisition and analysis by Mxpro-mx3000 (year 2007).
The identity of some PCR products was investigated by 2.5% agarose gel electrophoresis; to detect the characteristic band (251 base pair) of the 5′ un- translated region amplification.Citation10,Citation11
2.4 SYBR Green I based real time qualitative RT-PCR optimization
For optimization of the assay, we used the four standard HCV copy DNA molecules with known viral loads (provided with the HCV Qiagen kit TaqMan PCR Kit) which were processed by The SYBR Green I PCR described and standard curve was plotted.
The maximum sensitivity was obtained by testing various concentrations of primers. Optimal primer concentration was determined by using samples with different primer concentrations (0.1–0.5 µM). The amplification kit recruits TaqStart antibody that prevents primer extension at sub-stringent temperatures (HotStart) and minimize primer dimer formation.
2.5 Sensitivity and specificity
In order to determine the analytical sensitivity of our assay, RT-PCR experiments were performed on serial dilutions of standard copy DNA. To evaluate the specificity, known serum samples negative for HCV were extracted and tested.
3 Results
In this set of experiments, primer concentrations Ranging from 0.1 to 0.5 µM were tested. A primer concentration of 0.3 µM was selected since it produced maximum amplification without primer dimer formation. A MgCl2 concentration of 4 mM was used throughout the study.
The dynamic range of our assay covered four orders of HCV viral load from 102 to 106 IU/ml. HCV-specific fluorescent peaks were detected in the positive serum samples and standard panel dilution up to 50 IU/ml, but in none of the negative controls.
SYBR Green I fluorochrome binds to all amplicons without distinguishing between HCV and non-HCV products. Thus, the amplification product was characterized by melting curve analysis, wherein the HCV amplicon was easily distinguishable by its specific Tm value.
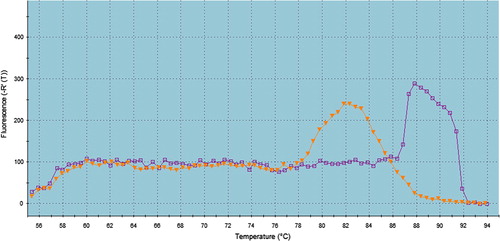
Under the employed experimental conditions, while analyzing the melting curve profiles of the PCR products, One of two different melting curve peaks either at 82 °C or 88 °C were observed, and both of them were associated with amplification plot ().
Fig. 1 Two cases detected positive by SYBR Green PCR method with two different melting curve peaks at 82 °C and 88 °C, respectively.
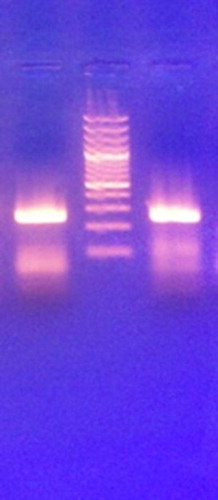
In the present study, conventional PCR targeting the 5́ UTR region was carried out to verify that these curves were related to the amplification of the specific target not the non-specific ones. The characteristic band of 251 bp was obtained for both curves indicating that the reason of obtaining two melting curves was due to different GC content with the same amplicon size ().
Fig. 2 Agarose gel electrophoresis showing the characteristic band (251 base pair) of 5′ untranslated region amplification in two cases positive by SYBR Green with two different melting curve peaks.
Out of the 220 chronic HCV patients included in this study, 138 patients (62.7%) were males and 82 (37.3%) were females. Regarding the associated risk factors, 77 cases (35%) had a history of hospital admission and 30 (13.6%) had a history of blood transfusion.
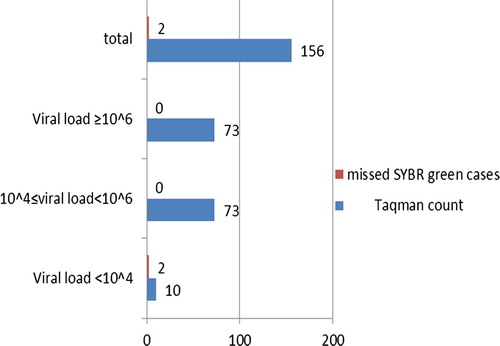
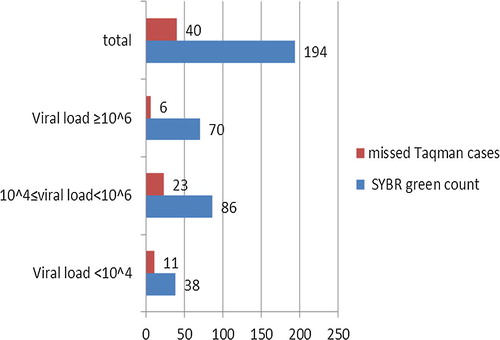
Among the 220 studied subjects, HCV-RNA was detected in 156 cases (70.9%) by TaqMan probe technique, while 194 cases (88.2%) were HCV-RNA positive by SYBR Green technique.
By both the techniques (TaqMan probe and SYBR Green), 154 (70%) were HCV-RNA positive and 24 (10.9%) were negative. On the other hand 40 patients (18.2%) were HCV-RNA positive by SYBR Green technique only and two (0.9%) by TaqMan probe only. Thus SYBR Green technique has a sensitivity of 79.38%, specificity of 92.31%, positive predictive value of 98.72% and negative predictive value of 37.50% (see –).
Fig. 3 Differences between both SYBR Green and TaqMan probe techniques regarding positivity and negativity.
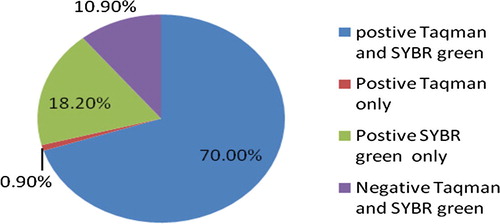
Table 1 Sensitivity, specificity and accuracy of SYBR green assay in relation of TaqMan probe as a gold standard technique.
By SYBR Green technique, 70 (31.8%) of the 220 chronic HCV patients showed a viral load equal or more than 106 IU/ml, 86 (39.1%) a viral load ranging from 104 to less than 106 IU/ml, and 38 (17.3%) with a viral load less than 104 IU/ml. On the other hand HCV-RNA was not detected in 26 (11.8%) patients
Among the 156 chronic HCV patients positive by TaqMan probe only 2 (1.3%) were negative by SYBR Green technique; in these two cases, the viral load was very low (less than 104 IU/ml).
4 Discussion
Egypt is facing a large burden of HCV disease, and prone to be the most affected nation of the world. In Egypt, the HCV prevalence and incidence across the diverse population groups are much higher than those in other countries.Citation12–Citation14 This makes HCV and the associated complications one of the leading public health concerns that Egypt has to cope.Citation15
The aim of this study was to compare between TaqMan probe technique and SYBR Green method in diagnosis and follow up of HCV patients. This study included 220 anti-HCV positive patients, 138 (62.7%) males and 82 (37.3%) females (male to female ratio: 1.75). In our patients, a history of blood transfusion was noted in 13.6% and hospital admission in 35%.
The conventional PCR techniques are relatively cumbersome, difficult to interpret, and prone to contamination.Citation16 Recently, real-time PCR has demonstrated its potential in detection of HCV RNA.Citation9,Citation16 The advantages of this technique over conventional PCR include higher sensitivity and specificity, accurate quantification, and most importantly very low possibility of cross-contamination.Citation17
SYBR Green is a fluorescent molecule that intercalates between double-stranded DNA molecules. It has many advantages over other real-time PCR detection methods, such as it is inexpensive and technically simple to use.Citation18 However, there is a major limitation in SYBR Green real-time PCR, as the dye indiscriminately binds to all double stranded DNA molecules resulting in non-specificity. In this case, both nonspecific and the amplified products are measured. Therefore, when SYBR Green is used, a melting-point analysis is performed to distinguish amplified products from others DNA molecules. The melting curve analysis identifies the amplified product depending on its length and GC content.Citation19
In the current study, the existence of two peaks should correspond to two amplified products. There are several situations wherein two peaks can appear in real-time PCR dissociation curve, e.g., non-specific amplifications, primer dimers and heterozygote polymorphism.Citation10 Conventional PCR targeting the 5́′ UTR region was carried out to verify the identity of these curves. The characteristic band of 251 bp was obtained for both curves indicating that the reason of obtaining two melting curves was due to different GC content with the same amplicon size The lower Tm belongs to a PCR product having a lower GC content.Citation11
Whereas, the labeled probes are expensive, add complexity to the oligonucleotide design, and affect the parameters involved in the classical amplification reaction.Citation18 PCR artifacts may be noted, especially when amplification cycles go above 30; besides this, the selected target sequence may also undergo mutations. Such mutations may render the target sequence undetectable by specific probes (i.e., TaqMan).Citation20 In SYBR Green-based methods such artifacts are easily recognized during melting curve analysis.Citation19
Similar to our results, Shahzamani et al. and Schmittgen et al.,Citation21,Citation22 reported a lower sensitivity of TaqMan probe technique compared with the use of the fluorescence dye, SYBR Green I.
In the present study only two out of 196 HCV-RNA positive cases were missed by SYBR Green Technique. Intermittent RNA positivity was observed in samples with low levels of virus; this could explain the two cases missed by SYBR Green technique.Citation23
During optimization of the assay, we focused on the compatibility of oligonucleotide pairs during amplification and on minimizing potential non-specific interactions such as primer dimers. In particular, the determination of primer concentrations played a pivotal role in enhancing the sensitivity and specificity. The SYBR Green I real-time RT-PCR assay depends on melting curve analysis to identify the target specifically. The Tm of the HCV melting curve was 82 °C with slight variations due to sequence variability between the positive patients.
One of the advantages of this assay is its low cost.
5 Conclusion
we reveal in this study that combining Syber green amplification (known with its high sensitivity) with melting curve analysis to increase the specificity of the assay.
Besides offering good level of sensitivity and specificity, relatively low cost, and low contamination rate, SYBR Green assay is perfectly suitable for both routine clinical use and large-scale screening of donated blood.
Large scale studies are needed for further evaluation of the SYBR Green assay for application as a screening assay for blood banking.
Conflict of interest
There are no conflict of interest in this article.
Notes
Peer review under responsibility of Alexandria University Faculty of Medicine.
Available online 24 November 2017
References
- WHO: Hepatitis C. World Health Organization Fact Sheet N °164 (updated April 2014). In: Media centre 2014. Available at: WHO Web site; <http://www.who.int/mediacentre/factsheets/fs164/en/>.
- D.LavanchyEvolving epidemiology of hepatitis C virusClin Microbiol Infect1722011107115
- P.P.ChouHepatitis C Virus: epidemiology, diagnosis, and patient managementLab Med.3820078691
- CDC Viral Hepatitis, Hepatitis C Information Page last updated: May 31, 2015 Available at: <http://www.cdc.gov/hepatitis/hcv/labtesting.htm>.
- C.L.J.GiachettiD.P.KolkJ.DockterHighly sensitive multiplex assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNAJ Clin Microbiol40200224082419
- J.L.GallardaBlood screening by nucleic acid amplification technology: current issues, future challengesMol Diagn520001122
- M.TajadiniM.PanjehpourS.H.JavanmardComparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypesAdv Biomed Res3201485
- J.D.ScottD.R.GretchMolecular diagnostics of hepatitis C virus infection: a systematic reviewJAMA2972007724732
- I.M.A.K.MackayA.NitscheReal-time PCR in virologyNucleic Acids Res30200212921305
- F.PonchelReal-time PCR using SYBR® GreenM.T.DorakReal-time PCR2007Taylor & FrancisNewcastle-upon-Tyne, UK
- H.Abtahi1M.R.SadeghiM.ShabaniCauses of bimodal melting curve: asymmetric guanine cytosine (GC) distribution causing two peaks in melting curve and affecting their shapesAfrican J Biotechnol1020111019610203
- W.SievertI.AltraifH.A.RazaviA systematic review of hepatitis C virus epidemiology in Asia, Australia and EgyptLiver Int3120116180
- M.J.AlterEpidemiology of hepatitis C virus infectionWorld J Gastroenterol13200724362441
- A.WasleyM.J.AlterEpidemiology of hepatitis C: geographic differences and temporal trendsSemin Liver Dis202000116
- Y.A.MohamoudG.R.MumtazS.RiomeD.MillerL.J.Abu-RaddadThe epidemiology of hepatitis C virus in Egypt: a systematic review and data synthesisBMC Infect Dis132013288309
- M.J.EspyJ.R.UhlL.M.SloanReal-time PCR in clinical microbiology: applications for routine laboratory testingClin Microbiol Rev192006165256
- F.WatzingerK.EbnerT.LionDetection and monitoring of virus infections by real-time PCRMol Aspects Med272006254298
- A.C.AldeaL.FolgueiraR.DelgadoJ.R.OteroRapid detection of herpes simplex virus DNA in genital ulcers by real-time PCR using SYBR Green I dye as the detection signalJ Clin Microbiol40200210601062
- C.NellåkerU.WållgrenH.KarlssonMolecular beacon-based temperature control and automated analyses for improved resolution of melting temperature analysis using SYBR I green chemistryClin Chem53200798103
- D.GibelliniF.VitoneP.SchiavoneG.FurliniM.C.ReSimultaneous detection of HCV and HIV-1 by SYBR Green real time multiplex RT-PCR technique in plasma samplesMol Cell Probes202006223229
- K.ShahzamaniF.SabahiS.MeratRapid Low-cost detection of Hepatitis C virus RNA in HCV infected Patients by real-time RT-PCR using SYBR Green IArch Iran Med142011396400
- T.D.SchmittgenB.A.ZakrajselA.G.MillsV.GornM.J.SingerM.W.ReedQuantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methodsJ Analyt Biochem2852000194204
- B.S.AnandM.VelezAssessment of correlation between serum titers of hepatitis C virus and severity of liver diseaseJ Gastroenterol10200424092411