?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Potassium ion is an essential macronutrient for plant growth and survival.
involves several biochemical and physiological pathways and hence its regulation is essential for normal metabolism and cellular functions.
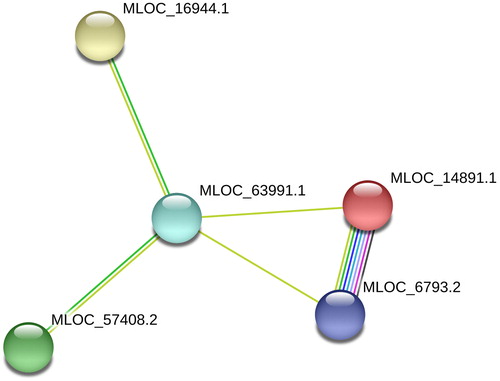
homeostasis is maintained by many proteins in a coordinated manner. In barley at least fifteen proteins are directly involved in potassium transport. Interaction among these proteins forms a network along with forty-nine other proteins. We retrieved the potassium transporter interaction network from STRING and the network was analyzed for centrality measures using Cytoscape. Based on the centrality measures such as betweenness, bridging, closeness, degree, eccentricity, eigenvector, radiality and stress, the proteins MLOC_6793.2, MLOC_14891.1, MLOC_16944.1, MLOC_57408.2, and MLOC_63991.1 are found to be most important for the network. These five proteins interact among themselves and form a subgraph. MLOC_63991.1 is the most essential protein as it links other 4 proteins in the network. MLOC_63991.1 is a monomer with 769 amino acid sequences. This protein is predicted and described as potassium transporter.
1 Introduction
Potassium is one of the vital plant nutrients. It is essential for maintaining electrical potential, hydrostatic pressure and biochemical activity for many enzymes Citation[1].
is transported by non-specific ion channels and high affinity transporter present in plasma membrane in the root system Citation[2]. These protein channels play a crucial role in maintaining ion homeostasis inside the cell Citation[3].
is transported by active transport against concentration gradient Citation[4].
A number of abiotic stresses affect the intercellular balance of and leads to the impairment in plant growth and reproduction. In barley (Hordeum vulgare) the existence of ion channels was stated by many researchers Citation[5,6]. The stress tolerant plant accessions try to maintain the normal and even high concentration of
during stressed condition. The background of the transport mechanism involved in the uptake and maintenance of higher concentration of
in plants has an agricultural significance Citation[5]. In this paper a computational approach is used to identify the role of potassium transporter in barley plant. We assume that the ion transporters present in barley forms an interaction network among them in a coordinated manner to regulate the uptake of
.
Materials and method
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) is a database meant for predicting the protein–protein interactions of known and predicted proteins based on experimental data, computational estimate methods and public text collections Citation[7]. A total of fifteen potassium transporters were identified till date in barley as mentioned in PFAM and INTERPRO protein domains. Potassium transporter expressed in barley plants were MLOC_10758.2, MLOC_11567.1, MLOC_11780.6, MLOC_16004.3, MLOC_16185.1, MLOC_30826.1, MLOC_37174.3, MLOC_53246.1, MLOC_58287.2, MLOC_62643.1, MLOC_63991.1, MLOC_66784.2, MLOC_70288.1, MLOC_71693.1, and MLOC_74565.1 (Table 1). All the fifteen proteins were analyzed by STRING Citation[6].
Table 1 Potassium transporters predicted in Hordium vulgare as per STRING data base.
These proteins have no direct interaction with each other. In order to find the relationship between these proteins, forty-nine 1st shell proteins were added. A total of sixty-four proteins formed a highly complex protein–protein interaction (PPI) network with 448 edges. The interaction may be physical or functional. These databases provide functional enhancements analysis report. The functional classification systems use in house prediction along with COG, Ensembl, Intact, RefSeq, PubMed, Reactome, DIP, BioGRID, MINT, KEGG, SGD, FlyBase, SwissProt/UniProt, SwissModel, HUGO, OMIM, NCI/Nature PID, PDB, The Interactive Fly, BioCyc, Gene Ontology, and SIMAP Citation[6]. This PPI network is highly useful to identify the cellular processes involved in potassium transport at system-level. Interaction networks can also suggest new directions for the experimental studies.
PPI network formed by 63 proteins and with 448 interactions were further analyzed using graph theory based Centrality measures. The software Cytoscape (v 3.6.1) helps us to visualize molecular interactions and to analyze networks of any kind. Its plugins help to analyze molecular profiles.
Result
The fifteen potassium transporter proteins had no direct or indirect interaction among them. These proteins interact with 49 other proteins and form an interaction network with the medium confidence level (0.4) for minimum required interaction score (Table 2)().
Fig. 1 Protein–protein interaction network formed by fifteen potassium transporters and its forty nine first shell interaction proteins.
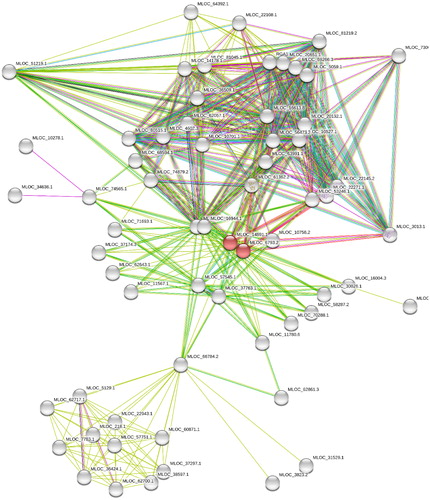
Table 2 Predicted functional partners for the Potassium transporter with medium level of confidence with first shell interaction based on gene neighborhood, text mining and experimentally determined scores.
The interactions are predicted from curated databases, experiments, gene neighbor-hood, and text mining. The interactions may also be predicted based on co-expression, and protein homology. For a similar kind of network the expected number of edges was 197. However, the actual number of edges is 448, which is much higher. An average node degree was 14.2, average local clustering coefficient was 0.622 and PPI enrichment -value was less than
. The network is given in .
PFAM protein domains for potassium transporters and its associated protein showed that 10 domains were significantly enriched by 36 proteins (Table 3). Function and domain for 27 proteins were not known. Table 4 represents the INTERPRO protein domains and features for potassium transporters and its associated protein. It showed that 25 domains were significantly enriched by 43 proteins and for 20 proteins function and domain are not known. It also predicts that 11 proteins are involved in more than one domain. Table 5 represents the average, minimum, and maximum score for the protein–protein interaction network analysis by STRING.
Table 3 PFAM protein domains for potassium transporters and its associated protein.
Table 4 INTERPRO protein domains and features for potassium transporters and its associated protein.
Table 5 The average, minimum and maximum score for the analysis of the protein–protein interaction network by STRING.
The centrality measures considered for protein–protein interaction network for potassium transporter in barley were Betweenness, Bridging, Closeness, Degree, Eccentricity, Eigenvector, Radiality, and Stress. The average, minimum and maximum values for all the centrality measurements for the node is presented in Table 6 for the network formed between the potassium transporters. The centrality measures for the top five proteins are listed in Table 7.
Table 6 Cytoscape analysis of protein–protein interaction network for the protein (node) and its centrality measurement.
Table 7 Top five proteins based on the centrality measures.
List 1. The proteins having above average values for the centrality measures
Betweenness
MLOC_6793.2; MLOC_14891.1; MLOC_57545.1; MLOC_37763.1; MLOC_16944.1; MLOC_57408.2;MLOC_74565.1; MLOC_63991.1; MLOC_16004.3; MLOC_66784.2
Bridging
MLOC_6793.2; MLOC_14891.1; MLOC_57545.1; MLOC_37763.1; MLOC_16944.1; MLOC_57408.2;MLOC_74879.2; MLOC_61362.2; MLOC_68594.1; MLOC_4602.3; MLOC_74565.1; MLOC_63991.1;MLOC_16004.3; MLOC_53246.1; MLOC_11780.6; MLOC_66784.2; and MLOC_62861.3
Closeness
MLOC_6793.2; MLOC_14891.1; MLOC_57545.1; MLOC_37763.1; MLOC_80515.1; MLOC_20132.1;MLOC_81219.2; MLOC_56473.2; MLOC_16613.8; MLOC_10527.1; MLOC_62057.1; MLOC_16944.1;MLOC_57408.2; MLOC_74879.2; MLOC_61362.2; MLOC_68594.1; MLOC_4602.3; MLOC_36509.1;MLOC_10701.1; MLOC_74565.1; MLOC_63991.1; MLOC_3013.1; MLOC_22271.1; MLOC_22145.2;MLOC_10758.2; MLOC_51219.1; MLOC_53246.1; and MLOC_66784.2
Degree
MLOC_6793.2; MLOC_14891.1; MLOC_57545.1; MLOC_37763.1; MLOC_80515.1; MLOC_20132.1;MLOC_81219.2; MLOC_56473.2; MLOC_16613.8; MLOC_10527.1; MLOC_81045.1; MLOC_62057.1;MLOC_14178.1; MLOC_16944.1; MLOC_57408.2; MLOC_4602.3; MLOC_36509.1; MLOC_10701.1;MLOC_69266.3; RGA1; MLOC_20651.1; MLOC_5059.1; MLOC_63991.1; MLOC_51219.1; MLOC_22108.1; MLOC_53246.1; and MLOC_66784.2
Eccentricity
MLOC_6793.2; MLOC_14891.1; MLOC_57545.1; MLOC_37763.1; MLOC_80515.1; MLOC_20132.1;MLOC_81219.2; MLOC_56473.2 MLOC_16613.8; MLOC_10527.1; MLOC_62057.1; MLOC_16944.1;MLOC_57408.2; MLOC_74879.2; MLOC_61362.2; MLOC_68594.1; MLOC_4602.3; MLOC_36509.1;MLOC_10701.1; MLOC_74565.1; MLOC_63991.1; MLOC_3013.1; MLOC_22271.1; MLOC_22145.2;MLOC_10758.2; MLOC_16004.3; MLOC_51219.1; MLOC_70288.1; MLOC_11567.1; MLOC_71693.1;MLOC_62643.1; MLOC_30826.1; MLOC_37174.3; MLOC_58287.2; MLOC_53246.1; MLOC_11780.6; andMLOC_66784.2
Eigenvector
MLOC_6793.2; MLOC_14891.1; MLOC_80515.1; MLOC_20132.1; MLOC_81219.2; MLOC_56473.2;MLOC_16613.8; MLOC_10527.1; MLOC_81045.1; MLOC_62057.1; MLOC_14178.1; MLOC_16944.1;MLOC_57408.2; MLOC_4602.3; MLOC_36509.1; MLOC_10701.1; MLOC_69266.3; RGA1; MLOC_20651.1; MLOC_5059.1; MLOC_63991.1; MLOC_3013.1; MLOC_22271.1; MLOC_22145.2; MLOC_51219.1;MLOC_22108.1; MLOC_53246.1; and MLOC_64392.1
Radiality
MLOC_6793.2; MLOC_14891.1; MLOC_57545.1; MLOC_37763.1; MLOC_80515.1; MLOC_20132.1;MLOC_81219.2; MLOC_56473.2; MLOC_16613.8; MLOC_10527.1; MLOC_62057.1; MLOC_16944.1;MLOC_57408.2; MLOC_74879.2; MLOC_61362.2; MLOC_68594.1; MLOC_4602.3; MLOC_36509.1;MLOC_10701.1; MLOC_74565.1; MLOC_63991.1; MLOC_3013.1; MLOC_22271.1; MLOC_22145.2;MLOC_10758.2; MLOC_51219.1; MLOC_71693.1; MLOC_53246.1; and MLOC_66784.2
Stress
MLOC_6793.2; MLOC_14891.1; MLOC_16944.1; MLOC_57408.2; MLOC_63991.1; MLOC_53246.1;and MLOC_66784.2
Discussion
The interactions in the living systems are usually depicted as networks. Network biology is a part of systems biology which enables us to understand the property of individual protein by analyzing the interactions between them via graph theory. Protein–protein interaction network is a mathematical expression of relationship between them. It has very high biological significance as it is specific Citation[8]. Based on centrality measures on the interaction between the potassium transporters in Hordeum vulgare (NCBI taxonomy Id is 4513) it was found that proteins MLOC_6793.2, MLOC_14891.1, MLOC_16944.1, MLOC_57408.2, and MLOC_63991.1 were the most essential for the network. Moreover these five proteins interact among them and forms a subgraph (). MLOC_63991.1 is found to be the most essential as it links other 4 proteins in the network.
The description of these proteins which are available in the literature are listed below:
MLOC_6793.2 has 482 amino acid sequences. Its molecular functions are orotate phosphoribosyl transferase activity and orotidine-5’-phosphate decarboxylase activity based on EMBL-EBI. It involves inasuridine-5’-phosphate biosynthesis pathway Citation[9]. MLOC_6793.2 is a predicted protein also known asMLOC_6793.4, F2D3N8, MLOC_6793.5, and MLOC_6793.2. MLOC_14891.1 is a predicted protein with 477 amino acid sequences [a.k.a. M0UW37, MLOC_14891,F2E520_HORVD, and MLOC_14891.1]. It exhibits orotate phosphor ribosyl transferase activity, orotidine-5’-phosphate decarboxylase activity, ’de novo’ pyrimidine nucleobase biosynthetic process, and ’de novo’ UMP biosynthetic process based on InterPro and gene ontology [Citation10]. MLOC_16944.1 is also known as M0V1P8_HORVD, AK356552, MLOC_16944, MLOC_16944.1. It was predicted as a functional partner of sodium/hydrogen exchanger. This predicted protein is made up of 535 amino acid sequences. MLOC_57408.2 is a Predicted protein with 1107 amino acid sequences [a.k.a. MLOC_57408.3, M0X6E4, and MLOC_57408.2]. It is a predicted functional partner of MLOC_30826.1, a potassium transporter [Citation11]. MLOC_63991.1 is a monomer with 769 amino acid sequences. This protein is predicted and described as potassium transporter [a.k.a. MLOC_63991.2, BAJ85390.1, F2DCE1, and MLOC_63991.1] [Citation10].
2 Conclusion
Analysis of proteins in crops has become fairly easy after the introduction of software such as Cytoscape, STRING, etc. Various cereal crops have been studied to improve the crop yield and nutritional value. Abiotic stresses such as salinity, drought, temperature, etc. alter the protein expression in plants. This study helps in identifying w proteins which are required to improve the plant tolerance to abiotic stresses.
References
- BrittoD.T.KronzuckerH.J., Cellular mechanisms of potassium transport in plants Physiol. Plant 1332008 637–650
- QiZ.HamptonC.R.ShinR.BarklaB.J.WhiteP.J.SchachtmanD.P., The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis J. Exp. Bot. 592008 595–607
- FulgenziF.R.PeraltaM.L.ManganoS.DannaC.H.VallejoA.J.PuigdomenechP.Santa-MaríaG.E., The ionic environment controls the contribution of the barley HvHAK1 transporter to potassium acquisition Plant Physiol. 472008 252–262
- MaathuisF.J.SandersD., Mechanisms of potassium absorption by higher plant roots Physiol. Plant 961996 158–168
- Nieves-CordonesM.AlemánF.MartínezV.RubioF., K+ Uptake in plant roots. The systems involved, their regulation and parallels in other organisms J. Plant Physiol. 171 9 2014 688–695
- SzklarczykD.MorrisJ.H.CookH.KuhnM.WyderS.SimonovicM.SantosA.DonchevaN.T.RothA.BorkP.JensenL.J.von MeringC., Quality-controlled protein-protein association networks, made broadly accessible The STRING database in 2017 Nucl. Acids Res. 452017 D362–68
- https://string-db.org, [Online]. Available:https://string-db.org/cgi/network.pl.
- SzklarczykD.MorrisJ.H.CookH.KuhnM.WyderS.SimonovicM.SantosA.DonchevaN.T.RothA.BorkP.JensenL.J.von MeringC., The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible Nucl. Acids Res. 452017 D362–68
- EMBL,www.ebi.ac.uk, EMBL-EBI 2018, Cambridgeshire, CB10 1SD, UK. 2018. [Online]. Available:https://www.ebi.ac.uk/training [Accessed 25 may 2018].
- www.arabidopsis.org, The Ohio State University-The Arabidopsis Information Resource (TAIR) [Online]. Available:https://www.arabidopsis.org/download_files/Pathways/Data_dumps/backslashPMN8_2013/barleycyc_pathways.20130709. [Accessed 7 NOV 2018].
- www.bioinfogenome.net, OrthoVenn, [Online]. Available:http://www.bioinfogenome.net/backslashOrthoVenn/cluster_info.php?ID=g5&overlapbinid=111111&clusterid=cluster8199. [Accessed 7 2018].