?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Microorganisms have an effective capability to degrade synthetic dyestuff enzymatically. In this study, two 16S rRNA identified Pseudomonas aeruginosa bacterial isolates (ASU3 and ASU6) have been screened to enzymatically degrade both Disperse Blue 64 (DB64) and Acid Yellow 17 (AY17) which resembles 2 of the synthetic dyes that causes aquatic environmental pollution. The expression of the azoR1 gene has been detected in the strains, the degradation percentage of DB64 and AY17 by stain ASU6 was found to exceed the ASU3. This returned to the differences in the expression levels of azoR1 between the ASU6 and the ASU3 which was confirmed by RT-PCR. In addition, the specific activity (U/mg protein) of azoreductase in ASU6 was higher than ASU3 when MethylRed (MR), DB64 and AY17 were used as a substrate, separately. The azoreductase kinetics was studied to find the optimum condition for maintaining the stability of the enzyme. The degraded products separated by HPLC/MS were tested for their toxicity on two crop plants and they were nontoxic to the plants.
1 Introduction
Most of the colored substances present in the environment and resistant to decomposition by light, water, chemicals and microbial activity are identified as synthetic dyes [Citation1,Citation2] . Azodyes are a large group of aromatic compounds that consist of one or more chromophoric azo group (–N=N) that are present in the water body due to many industrial activities and causes aquatic environmental pollutants. Disperse blue (DB64) is an anthraquinone dye represent the second most important class of commercial dyes which mainly used for dyeing wool, polyamide and leather [Citation3]. Acid Yellow 17 (AY17) is a commercial organic azodye that have a genotoxic effect [Citation4]. They cause skin irritation, allergy [Citation5], mutagenicity and carcinogenicity [Citation6] in humans. Many strategies have been developed to remove the industrial dyes from the water body, such as chemical oxidation and reduction [Citation7], adsorption [Citation8] and membrane filtration [Citation9], photo-catalytic degradation [Citation10], coagulation and precipitation [Citation11], electrocogulation [Citation12], photoelectrocatalysis [Citation13] and electrochemical oxidation [Citation14]. Unfortunately, these strategies are expensive and may produce toxic products [Citation2]. On the other hand, Using of microorganisms to remove these dyes are fast [Citation15,Citation16] , inexpensive, and the end products are safe [Citation17].
Different microorganisms, including bacteria [Citation18Citation[19]–Citation20] , fungi [Citation21,Citation22] algae [Citation23,Citation24] , are able to remove different type dye stuff. The removal of the dyes is carried out by different mechanisms. Biosorption on the cell wall due to the presence of different functional groups is one way to remove the dyes [Citation16,Citation25] . Another way is the enzymatically microbial degradation by the azodye–catabolic enzymes like azoreductase [Citation26], laccase [Citation27] and peroxidase [Citation28]. Azoreductases of microorganisms are favorable for the development of biodegradation systems for azo dyes, because these enzymes catalyze reductive cleavage of azo groups (–N=N–) under mild conditions [Citation29]. However, the aerobic treatment of azodyes requires a special aerobic azoreductase system which catalyzes the NADH or NAD(P)H-dependent reduction of azo compounds to the corresponding amines [Citation30]. This enzyme could not only reduce the carboxylic bacterial substrate but also it could reduce the sulfonated analogoes. A small number of bacterial species have been reported to degrade azodyes under aerobic condition such as Bacillus sp. OY1-2 [Citation31]. Pseudomonas [Citation32], Xenophilusazovorans [Citation33], and Sphingomonas sp. [Citation34]. This study aims to: (i) isolate and identify the azodyes- degrading Pseudomonas species, (ii) detect the azoR1 gene in isolated strains (iii) compare azoreductase activity, kinetics and gene expression at the RNA level.
2 Materials and methods
2.1 Strains isolation
The isolation of the dye decolorizing bacterial species was carried out according to Hesham et al. [Citation35] with some modifications. About 10 mL or mg of water effluent or sludge sample, respectively were suspended in 90 ml (MBS) medium. The medium was supplemented with 100 mg l−1 of DB64 or AY17 and 0.1% of glucose as a carbon source. The flasks were incubated on orbital shaker at 150 rpm at 30 °C. After 7 days incubation, an aliquot of 10% enriched cultures were transferred to a new flasks containing 90 ml fresh autoclaved MBS medium supplemented with previously mentioned dyes. This step was repeated five times to attain well-adapted dye decolorizing microorganisms.
To select Pseudomonas dye degrading strains, cultures were transferred to a King A agar medium supplemented with 100 mg l−1 of dye at pH 7.0 and 30 °C. Pseudomonas colonies were individually selected, purified and evaluated for their dye decolorizing ability.
2.2 Identification of azodye–degrading Pseudomonas spp
Colony-PCR was performed to amplify the 16S rRNA gene using the 27F (5-AGAGTTTGATCCTGGCTCAG-3) and 1492R (5-CGGCTACCTTGTTACGACTT-3) primers [Citation36]. PCR was programmed as following: 5 min at 95 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C and then one cycle of final extension at 72 °C for 10 min. PCR product was then analyzed on 1.5% agarose gel electrophoresis and then sequenced in both directions. The obtained sequences were aligned and compared using the BLAST search tool at the (http://www.pseudomonas.com/). To determine the taxonomic position of the isolate, a phylogenetic tree was constructed by MEGA version 4.0 software.
2.3 Measuring azodye degradation by the UV-vis Spectrophotometer
An overnight culture were inoculated in MBS medium supplemented with 0.1% of glucose and 100 mg l−1of the tested dyes at 30 °C under static condition. The percentage of decolorization was estimated from the following equation [Citation37]:(1)
(1) where A0; is the initial absorbance of dyes and At is the absorbance of dye at (t) time. The blank sample is the cultivation medium.
For determination the concentration of dye in the medium, standard curves of DB64 and AY17 with different concentrations (5–300 mg l−1) were made.
2.4 Crude enzyme extraction
Strains was grown in MBS liquid medium amended with 0.1% glucose and 100 mg l−1of methyl red to enhance the production of azoreductase enzyme. The cells were harvested by centrifugation and suspended in potassium phosphate buffer (50 mM, pH 7.4). The cells were crushed by sonication at 35 amplitude for 2 min (30-s interval) at 4 °C. The homogenate was then centrifuged at 5000 rpm for 30 min under cooling and the supernatant was transferred to a new tube and the pellet was removed. The total protein concentration was determined by Bradford method [Citation38], with bovine serum albumin as standard.
2.5 Azoreductase specific activity and substrate specificity
The azoreductase specific activity was assessed by the reduction of methyl red (MR) as the main substrate and oxidation of NADH using a modified procedure described by Zhao et al. [Citation39]. An enzyme assays mixture was prepared by mixing 600 µL phosphate buffer (50 mM and pH 7.5), 100 µL of dye substrate (50 µM), 100 µL of the crude enzyme and 200 µL of NADH (200 µM). MR, DB64 and AY17 were used as a specific substrate to determine the specific azoreductase activity and detect substrate specificity.
One unit of azoreductase activity was defined as one microgram of methyl reduced protein per minute per milligram. All enzyme assays were run in triplicate.
2.6 Factors affecting on azoreductase stability
Different pHs and temperatures were tested to determine their effect on the azoreductase stability. To determine the pH effect, the NADH and MR buffers was prepared with a range of different pH (from pH 5 to 9), while the acetate buffer was pH 5.5, the phosphate buffer was pH (6.5–7.5) and Tris-HCl buffer was pH [Citation8–Citation9] .
The temperature effect was determined by using variable temperature values ranged from 20 to 60 °C during the measurement with the UV-Vis Spectrophotometer. The temperature of all enzymes reaction mixture was adjusted to the desired temperature except the NADH.
2.7 Azoreductase kinetics
Different concentrations of MR (ranged from 17 to 3500 mM) were prepared to determine the azoreductase kinetics. All other constituents of reaction mixture were constant. Kinetic values such as Michaelis–Menten constant (K max) and maximal velocity (Vmax) were calculated from Lineweaver–Burk double reciprocal plots [Citation40].
2.8 The expression of the azoR1 gene
Reverse transcriptase PCR (RT-PCR) technique was used to determine the azoR1 gene expression. RNA was extracted by using SV Total RNA Isolation System (Promega). First-strand cDNA was reverse transcribed from one microgram of total RNA by using First-strand cDNA Synthesis Kit (Amersham Biosciences, GE Healthcare). RT-PCR was performed using (azoR1Fw: 5′-AACAGCGTTCGCTGGCGATG-3′ and azoR11Rv: 5′- AGGGATCGGCGTGGTTCATC-3′) primers which were designed to specifically amplify a 303 bp sequence in the azoR1 gene. PCR conditions were: initial denaturation for 5 min at 95 °C, 36 cycles with 1 min at 94 °C, 40 s at 56 °C, 60 s at 72 °C, and final extension for 10 min at 72 °C. The reaction products were analyzed on 1% agarose gel electrophoresis. The intensity of the bands was analyzed using Gene Profiler 4.03 computer software to determine the OD of each band and consequently calculate the concentration of DNA in the band.
2.9 Toxicity of metabolic products
The toxicity of the dyes and its metabolic products were determined by acute toxicity test on 2 popular crop plants in Egypt; monocot seed (Triticum aestivum) and dicot seed (Vicia faba). Briefly, the dyes degraded broths were centrifuged at 10000 rpm for 20 min under cooling. The supernatant was collected and sterilized by filtering. About 10 plant seeds were germinated in sterilized Petri dish and irrigated with dye filtrate (10 mL) at 30 °C for 7 days. Then the germination percentage, the root and shoot length were calculated. A positive and a negative control were included in the experiment. The positive was irrigated with the dyes buffers without the bacterial treatment while the negative control was irrigated with sterilized distilled water.
2.10 Determination of the metabolic products by LC-MS
10 mL of the metabolites were extracted with three equal volumes of Ethyl Acetate. The residual extracts were dried over anhydrous Sodium Sulfate and evaporated with rotary evaporator at 40 °C to 10 ml. The samples were dried in vacuum and stored at −20 °C until used. The metabolic products were analyzed by Waters Alliance Acquity UPLC - Xevo G2S-QTof instrument. Extracts were dissolved in 1 ml of 1:1 MeCN:H2O. 10 µl aliquot was taken and diluted with 90 µl 1:1 MeCN: H2O. 2 µl was injected for LC-MS analysis. The LC conditions: A = H2O + 0.1% HCOOH; B = MeCN + 0.1% HCOOH. 5% B–95% B: 0–12 min 95% B: 12–17 min 5% B: 17–20 min. The flow rate was 0.2 ml/min, column temperature was 40 °C and the Acquity UPLC BEH C18 column, 1.7 µm, 2.1 × 50 mm.
The MS conditions: Range: 100–1200 Da, Capillary: 3.5 V, Sampling cone: 40 V, Source temperature: 120 °C, Desolvation temperature: 200 °C, Cone gas: 25 L/hr, Desolvation: 600 L/hr and MSe: ramp collision energy from 15 to 45 V. The MS detector was performed by electrospray ionization in positive mode [ESI+].
3 Results and discussion
3.1 Azodye–degrading Pseudomonas spp isolation and identification
Twenty Pseudomonas strains were isolated from different textile dye effluent and screened for azodye degrading capability (results are not shown). Two strains ASU3 and ASU6 exhibited hyber azodye degrading capability. The strains were identified based on some morphological and biochemical activity using Bergey’s Manual of Systematic Bacteriology as Pseudomonas aeruginosa. The cell shape was short rods and gram-negative. They showed high growth on the different Pseudomonas selective media like Cetrimide, King A and King B and produce green pyocyanin pigment. They are oxidase and catalase positive strains and can oxidize glucose.
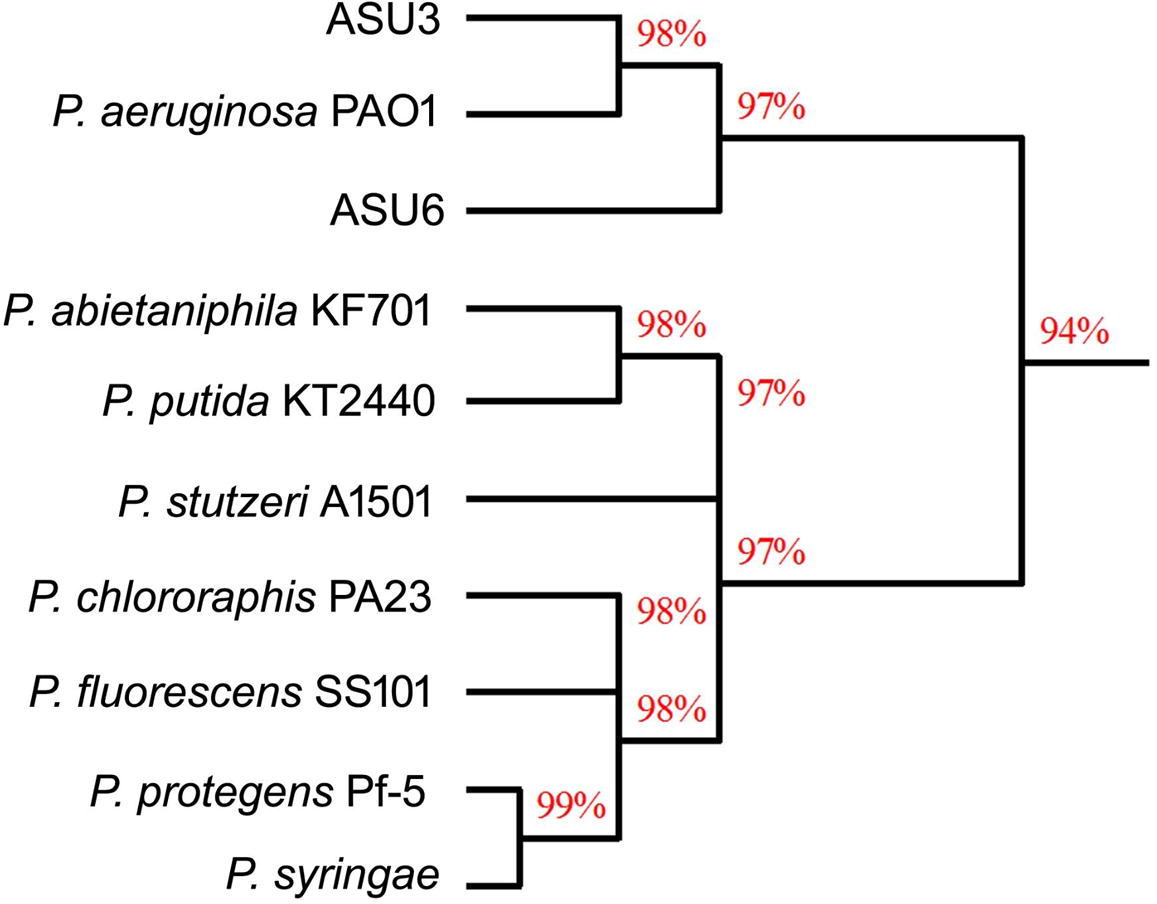
Moreover, we used the 16S rRNA gene sequencing to molecularly identify the ASU3 and ASU6 strains. The 16S rRNA identifications is more robust, reproducible, accurate and less subjective test results [Citation1,Citation41] . Based on the Blast results and the dendrogram demonstrating the relationship among some Pseudomonas strains (), ASU6 and ASU3 strains were highly similar to Pseudomonas aeruginosa with a similarity of 97–98%, respectively.
Fig. 1 Dendrogram demonstrating the relationship among the isolated strains (ASU3 and ASU6) and some Pseudomonas strains.
3.1.1 Degradation of azodyes
The capacity of Pseudomonas strains for the decolorization of various dyes has been studied in detail [Citation42] who reported that, the azo dyes (Acid Violet 7, Acid Red 151 and Reactive Black 5) were degraded at higher extent (>90%) than Indigo Carmin, Acid Red 183 (chromium complex) and anthraquinones (Reactive Blue 2 and Acid Green 27). Decolorization of waste waters containing reactive azo dyes was achieved by a culture of the bacteria P. luteola that reduced the azo bond in Red G [Citation43].
Using the UV-Vis Spectrophotometer, the degradation percentage of each dye was estimated for the two selected strains at the maximum absorbance (λmax) using equation [Citation1]. DB64 and AY17 were monitored at 600, and 398 nm, respectively. Strains ASU3 and ASU6 could degrade 61.2 and 96.8% of DB64 within 48 h and under static condition, respectively. At the same time, they could remove 38.6 and 91.2% of AY17, respectively ().
Fig. 2 Percentage of DB64 and AY17 degradation by strains ASU3 and ASU6 at pH 7.0, 30 °C and 100 mg l−1dye under static condition.
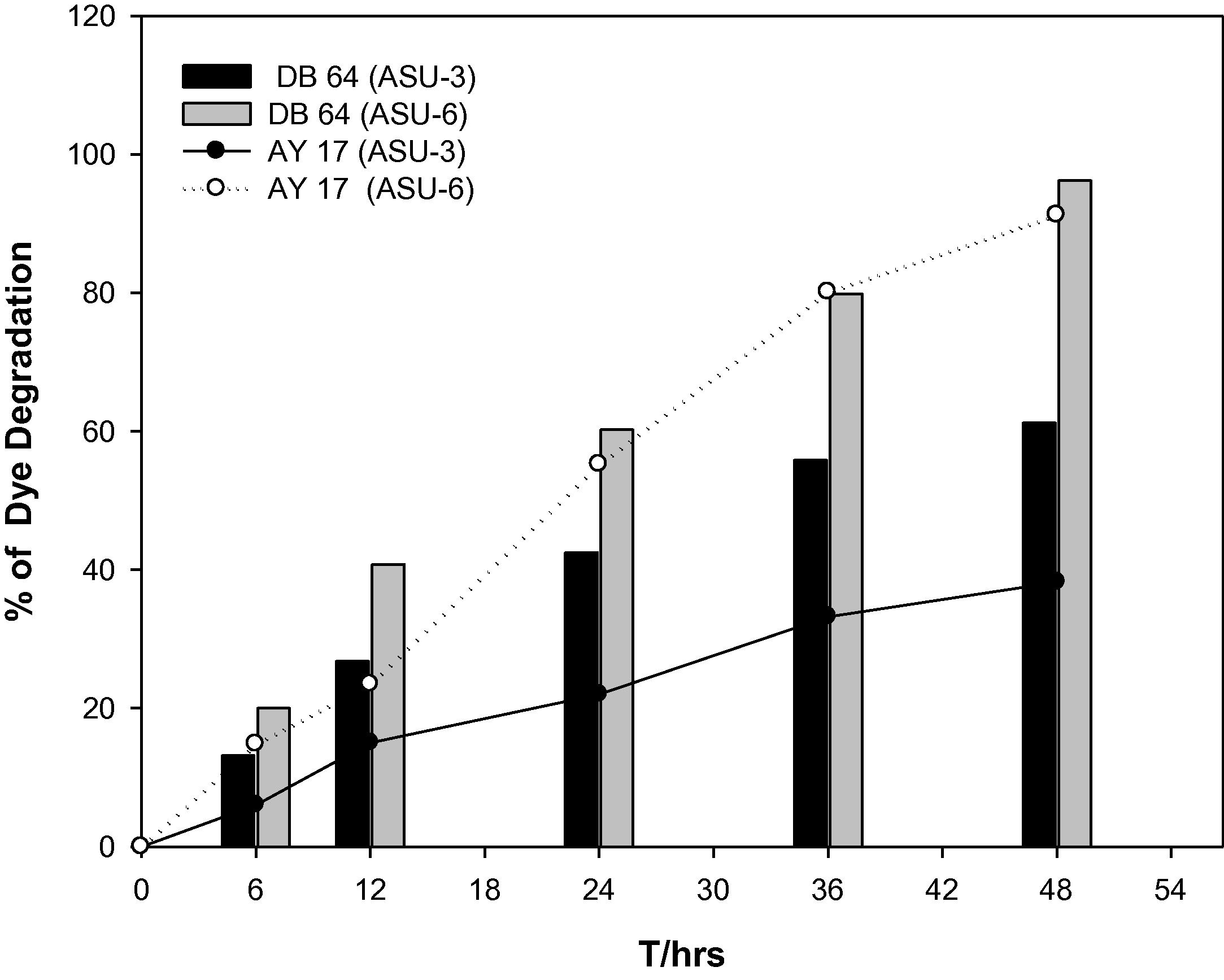
The results showed that the efficiency of two strains to degrade DB64 was higher than AY17. In addition, the ability of the strain ASU6 to degrade the two distinct types of azodyes exceeded the ability of ASU3.
3.1.2 azoR1 gene expression in isolated strains
To evaluate the effects of different substrate on the transcription of azoreductase gene, cells were cultured in media supplemented with 100mg l-1of either DB64, AY17 or MR for 48 h. RNA was directly extracted from cultivated strains after the decolorization of dye using the SV total RNA Isolation system (Promega). The expression of azoR1 gene was used to detect the activity of the ASU3 and ASU6 strains on the azodyes.
The transcription of azoR1 gene was variable from strain to another and from substrate to other. In this study, the expression of azoR1 gene in strain ASU6 was higher than its expression in ASU3 when they consumed MR and AY17 and the opposite occurred when the DB64 was consumed. When using AY17 as the main substrate, the transcription of azoR1 gene was significantly weak in the strain ASU6 with lnt OD 1.82 and band concentration was about 35.1 ng but it was hardly detected in ASU3; the band concentration was about 12.5 ng ( and ).
Fig. 3 Agarose electrophoresis of the RT-PCR product to test the expression of the azoR1 gene in both ASU6 and ASU3 isolates growing in different substrates (AY17, MR, and DB64).
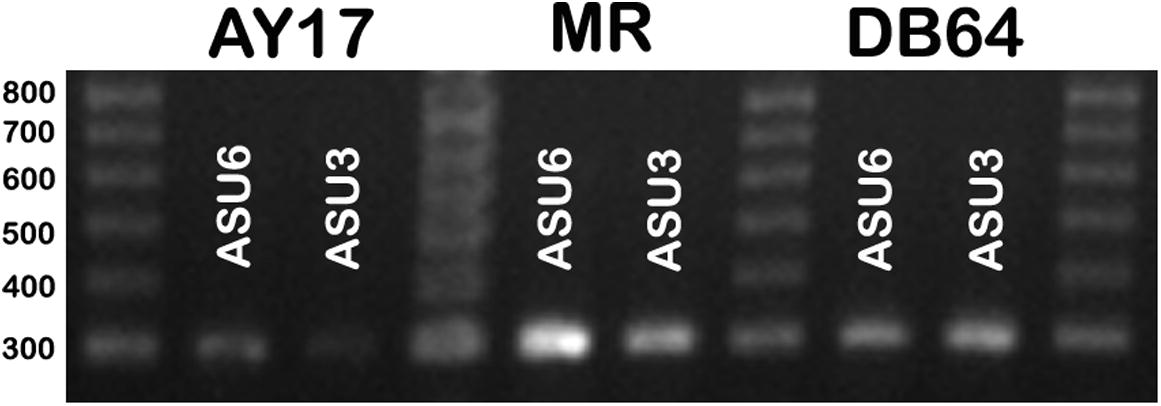
Table 1 The data of lnt OD and band concentration (ng) that extracted from gene profiler software.
Regarding the DB64-containing medium, the two strain exhibit a bright band. Interestingly, the gene expression as well as the band concentration in the strain ASU3 (93.3 ng) was more than ASU6 (63.97 ng). Moreover, the highest expression was detected when the strains were separately cultivated on the medium supplemented with MR (212.07 and 98.8 ng for ASU6 and ASU3, respectively) ( and ).
In addition, the transcription of azoR1 seemed to be subjected to the type and chemistry of the dye. The results of gene expression were highly supported the results of azoreductase specific activity and the degradation percentage except with the degradation of DB64. When the DB64 was used as a substrate, the RNA expression of azoR1 gene in strain ASU6 was lower than ASU3. However, the enzyme specific activity and the degradation percentage were higher. The reason of this phenomena may be due to the presence of other co-enzymes or cofactors cooperated to enhance the activity of azoreductase as well as the degradation of the DB64 in strain ASU6 rather than azoR1 encoding azoreductase as mentioned by Equation(1)(1)
(1) .
Leelakriangsak et al. [Citation44] reported the increase in the expression of genes (ppazoR1, ppazoR2 and ppazoR3) on the mRNA levels in P. aeruginosa. Töwe et al. [Citation45] also observed more significant increases in azoR1 and azoR2 gene expression in Bacillus subtilis in the presence of quinines.
3.2 Determination azoreductase specific activity and substrate specificity
Azoreductase is a key enzyme catalyzes the reduction of azodyes in versatile microorganisms. This enzyme could be inhibited by oxygen [Citation46]. The specific activity of azoreductase enzyme (U/ mg protein) was monitored for the two strains during the growth of strains on methyl red (MR), DB64 or AY17 as specific substrates in the presence of NADH as the electron donor (oxidative mediators). The experiment was carried out under a static condition at 30 °C. The maximum specific activity of ASU3 and ASU6 on different substrates was summarized in . The strains recorded highest values of azoreductase specific activity when the MR dye was applied as a specific substrate (0.92 and 0.3 U/mg protein for ASU6 and ASU3, respectively). On the other hand, the specific activity of azoreductase enzyme was at the lowest value when the AY17 was used. It was observed that, the specific activity of azoreductase producing strain ASU6 higher than strain ASU3. Thus, it could be said that, the specific substrate that will be applied to find the enzyme kinetics was MR.
Table 2 The aerobic azoreductase specific activity (U/ mg protein) of two strains on different substrates at 30 °C, pH 7.0, 50 µM substrate concentration and 200 µM NADH.
Due to a competition for consumption of NADH by aerobic respiration, which make the electron transfer from NADH to oxygen in order to form ATP [Citation47]. Many bacteria possess a specific cytoplasmic azoreductases system that catalyzed the electron transfer from NADH (electron donor) to azo bonds. So, the presence of NADH is very significant in the reduction of azodyes by aerobic bacteria [Citation46]. Thus, it could be concluded that, the aerobic azoreductase enzyme is an inducible enzyme required redox mediator for perfect performance. Likewise, that the presence of dyes as substrates stimulate the induction of this enzyme. Azoreductase activity in azodyes decolorizing bacteria has been extensively examined to elucidate azo dye reduction mechanism [Citation30,Citation39,Citation44,Citation48] .
3.3 Factors affecting azoreductase activity
Azoreductase reaction should take place at optimum condition. The most important condition affecting on the activity of the enzymes is pH and temperature. Hence, it is rattling important to determine the optimal conditions for the operation of enzymes. The activity of azoreductase was maximum at pH 7.5 for ASU3 and ASU6 at 30 °C (a). Thus, the optimal pH value for azoreductase enzyme was pH 7.5 when MR was used as a specific substrate. While the optimum temperature for azoreductase was at 40 °C for the two strains (b). No significant activity has been observed at 60 °C for strain ASU6. These came in context with many literatures which study the optimum condition for azoreductase [Citation46,Citation47] .
3.4 Kinetics of azoreductase
Using a different concentration of the substrate (MR) is a main factor to determine the enzyme kinetics. The kinetics parameters like Vmax (maximum velocity) and Km (Michaelis constant) were estimated and the velocity of azoreductase in ASU6 (10.6 µM/min/mg) was higher than ASU3 (0.5 µM/min/mg). However, the value Km were 125 and 3240 µM. The low value of km indicated that the affinity of substrate to azoreductase in ASU3 was higher than its affinity in ASU6 ().
Fig. 5 A Lineweaver-Burk plot for determination the kinetic parameters of azoreductase in ASU3 and ASU6 at pH 7.0 and temperature 40 °C.
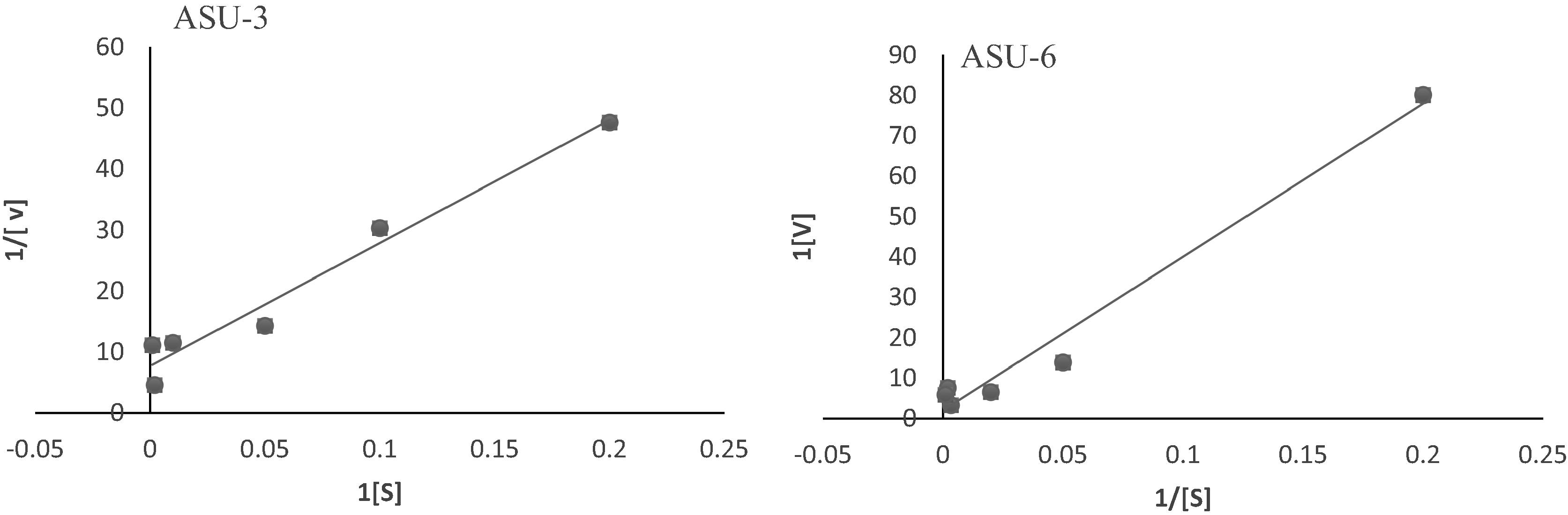
Many literatures reported that azoreductase enzyme is not only capable of reduction of azo dyes, but also it could reduce several naphtho-, benzo-, and anthraquinone compounds. In some cases, these compounds act as a better substrate than methyl red due to higher value of Km and Vmax than MR [Citation49]. The enzyme kinetics in two isolated strains indicated that the velocity Vmax and Km of the enzyme in ASU6 was higher than ASU3. Nevertheless, an enzyme has a low value of km it achieves its maximum catalytic efficiency at low substrate concentrations. ASU3 azoreductase was more effective as a catalyst than ASU6.
3.5 Phytotoxicity test
The use of untreated or treated wastewater contaminated with dyes in the land may have a harmful effect on the soil fertility and productivity. Thus, it was important to consider the effect of dyes and its product on two different crop plants. Because the strain ASU6 exhibited higher degradation ability to synthetic dyes than ASU3, it was selected for phytotoxicity studies. The results demonstrated that, the percentages of germination were significantly low when the seeds treated with the crude dyes compared to that treated with the metabolic products. The highest germination percentage achieved when the DB64 metabolic products were applied on monocot and dicot seeds 87% and 80%, respectively (). The length of shoot and root was also significantly affected.
Table 3 The phytotoxicity test on Triricum endobyoticum and Vicia faba plant rowing on the tested dyes and its metabolic products.
The results showed that, the toxicity of dyes and its degradation products on the plant was different according the type and chemistry of these compounds. The effective microbial degradation of dyes will lead to safe metabolic products.
3.6 Metabolic products by HPLC–MS
The metabolic products that produced from the degradation of DB64 by strain ASU6 was analyzed by HPLC–MS for tentative identification of intermediate compounds. The HPLC–MS chromatogram showed that, there were many tentative products produces after incubation of dye with strain ASU6. Four tentative compounds have a good match with the structure of DB64 that were listed in . These compounds indicated that the main behavior of the strain ASU6 for degradation of DB64 was the reduction of anthraquinone ring.
Table 4 Intermediate metabolites identified by HPLC/MS.
The tentative products gave an impression that, there were two expected metabolic pathways () for degradation of DB64. The first one was that the DB64 can undergo reduction through azoreductase via two electrons transfer that lead to formation of hydroquinone (; No. 4). The subsequent reduction mechanism finally leads to ring cleavage (; No. 5). The second pathway was to eliminate the methoxy groups from the dye (; No. 2) and subsequently one of the amino benzene was liberated and the other was hydrated (; No. 3).
The mechanism of hyrdoquinone formation was that NADH acted as redox mediators which were enzymatically reduced by the cells to form hydroquinones, which in turn reduced the azo dye. The quinine reductase activity was observed in the cell membrane and hence the transport of the hydroquinone /quinine redox mediator and the highly polar sulfonated azo dye across the cell membrane was not required.
The metabolites could be detected from the degradation of AY 17( & ) were only two metabolites ( No; 7&8) indicating the cleavage of diazo bond and formation of sulfonated aromatic amines.
4 Conclusion
The percentage of degradation of many dyes is controlled by the type and chemistry of dye and the ability of bacteria to produce the azoreductase enzyme. The percentage of dye degradation was variable within the two isolated strains why the strain ASU3 and ASU6 could degrade anthraquinone dye DB64 with higher efficiency than AY17. May be due to the anthraquinone dyes are easier to degrade than azodyes one or the presence complex structure of AY17 make obstacle its degradation. Other causes may be due to the bacterial azoR1gene expressions in the presence of DB64 was higher than AY17.
Azoreductase is the main enzyme responsible for degradation of DB64 and AY17. When the transcription of azoR1 gene decreases, it led to decrease the azoreductase activity and its velocity toward the substrate and consequently, depletion in the degradation rate. Along the DB64 degradation process by ASU6, four metabolites can be detected that indicated the reduction of the anthraquinone ring by azoreductase. These metabolites were proved to be non-toxic to the crop plants.
Acknowledgments
A great acknowledge to the Assiut University Mycological Center (AUMC) for helping in the identification and preservation of samples in (AUMC) culture collection. Also, the authors thank Prof. Dr. Mostafa Fouad, and Dr. Usama Ramadan, Faculty of Pharmacy, El Minia University, Egypt for their help in the HPLC/MS analysis and identifications.
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- M.LeelakriangsakMolecular approaches for bacterial azoreductasesSongklanakarin J Sci Technol3562013647657
- P.SinghL.IyengarA.PandeyBacterial decolorization and degradation of azo dyes. Microbial degradation of xenobiotics2012Springer101133
- A.ReifeH.S.FreemanCarbon adsorption of dyes and selected intermediatesEnviron Chem Dyes Pigm1996331
- H.B.MansourD.CorrolerD.BarillierK.GhediraL.ChekirR.MosratiEvaluation of genotoxicity and pro-oxidant effect of the azo dyes: acids yellow 17, violet 7 and orange 52, and of their degradation products by Pseudomonas putida mt-2Food Chem Toxicol459200716701677
- J.J.SorianoJ.Mathieu-DenoncourtG.NormanS.R.de SollaV.S.LangloisToxicity of the azo dyes Acid Red 97 and Bismarck Brown Y to Western clawed frog (Silurana tropicalis)Environ Sci Pollut Res215201435823591
- C.WangA.YedilerD.LienertZ.WangA.KettrupOzonation of an azo dye CI Remazol Black 5 and toxicological assessment of its oxidation productsChemosphere527200312251232
- X.FlorenzaA.M.S.SolanoF.CentellasC.A.Martínez-HuitleE.BrillasS.Garcia-SeguraDegradation of the azo dye Acid Red 1 by anodic oxidation and indirect electrochemical processes based on Fenton's reaction chemistry. Relationship between decolorization, mineralization and productsElectrochim Acta1422014276288
- V.GuptaB.GuptaA.RastogiS.AgarwalA.NayakA comparative investigation on adsorption performances of mesoporous activated carbon prepared from waste rubber tire and activated carbon for a hazardous azo dye—Acid Blue 113J Hazard Mater18612011891901
- A.MittalV.ThakurV.GajbeEvaluation of adsorption characteristics of an anionic azo dye Brilliant Yellow onto hen feathers in aqueous solutionsEnviron Sci Pollut Res196201224382447
- Y.CaiM.StrommeA.MelhusH.EngqvistK.WelchPhotocatalytic inactivation of biofilms on bioactive dental adhesivesJ Biomed Mater Res B Appl Biomater102120146267
- A.K.VermaR.R.DashP.BhuniaA review on chemical coagulation/flocculation technologies for removal of colour from textile wastewatersJ Environ Manage9312012154168
- A.S.FajardoR.F.RodriguesR.C.MartinsL.M.CastroR.M.Quinta-FerreiraPhenolic wastewaters treatment by electrocoagulation process using Zn anodeChem Eng J2752015331341
- S.Garcia-SeguraE.BrillasApplied photoelectrocatalysis on the degradation of organic pollutants in wastewatersJ Photochem Photobiol C: Photochem Rev2017
- A.M.S.SolanoS.Garcia-SeguraC.A.Martínez-HuitleE.BrillasDegradation of acidic aqueous solutions of the diazo dye Congo Red by photo-assisted electrochemical processes based on Fenton’s reaction chemistryAppl Catal B1682015559571
- A.KhalidM.ArshadD.E.CrowleyAccelerated decolorization of structurally different azo dyes by newly isolated bacterial strainsAppl Microbiol Biotechnol7822008361369
- M.SolísA.SolísH.I.PérezN.ManjarrezM.FloresMicrobial decolouration of azo dyes: a reviewProcess Biochem4712201217231748
- C.J.OgugbueT.SawidisBioremediation and detoxification of synthetic wastewater containing triarylmethane dyes by Aeromonas hydrophila isolated from industrial effluentBiotechnol Res Int20112011
- F.ElisangelaZ.AndreaD.G.FabioCristiano R.de MenezesD.L.ReginaC.-P.ArturBiodegradation of textile azo dyes by a facultative Staphylococcus arlettae strain VN-11 using a sequential microaerophilic/aerobic processInt Biodeterior Biodegradation6332009280288
- G.GhodakeS.JadhavV.DawkarS.GovindwarBiodegradation of diazo dye Direct brown MR by Acinetobacter calcoaceticus NCIM 2890Int Biodeterior Biodegradation6342009433439
- R.SarataleG.SarataleD.KalyaniJ.-S.ChangS.GovindwarEnhanced decolorization and biodegradation of textile azo dye Scarlet R by using developed microbial consortium-GRBioresour Technol1009200924932500
- N.S.MauryaA.K.MittalP.CornelE.RotherBiosorption of dyes using dead macro fungi: effect of dye structure, ionic strength and pHBioresour Technol9732006512521
- P.KaushikA.MalikFungal dye decolourization: recent advances and future potentialEnviron Int3512009127141
- H.H.OmarAlgal decolorization and degradation of monoazo and diazo dyesPak J Biol Sci1110200813101316
- E.DaneshvarM.KoushaM.S.SohrabiA.KhataeeA.ConvertiBiosorption of three acid dyes by the brown macroalga Stoechospermum marginatum: isotherm, kinetic and thermodynamic studiesChem Eng J1952012297306
- A.M.MawadN.M.YousefA.A.ShoreitBioremediation of Acid blue 25 dye by anthracene degrading Pseudomonas pseudoalcaligenes ASU-016Catrina Int J Environ Sci1020155360
- Puvaneswari N, Muthukrishnan J, Gunasekaran P. Toxicity assessment and microbial degradation of azo dyes. 2006.
- S.-S.MirzadehS.-M.KhezriS.RezaeiH.ForootanfarA.H.MahviM.A.FaramarziDecolorization of two synthetic dyes using the purified laccase of Paraconiothyrium variabile immobilized on porous silica beadsJ Environ Health Sci Eng12120146
- S.SarojK.KumarM.PrasadR.SinghDifferential expression of peroxidase and ABC transporter as the key regulatory components for degradation of azo dyes by Penicillium oxalicum SAR-3Funct Integr Genomics1442014631642
- M.NakanishiC.YatomeN.IshidaY.KitadePutative ACP phosphodiesterase gene (acpD) encodes an azoreductaseJ Biol Chem2764920014639446399
- H.ChenS.L.HopperC.E.CernigliaBiochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric NADPH-dependent flavoproteinMicrobiology151Pt 5200514331441
- Y.SuzukiT.YodaA.RuhulW.SugiuraMolecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1-2 isolated from soilJ Biol Chem27612200190599065
- A.A.TelkeS.M.JoshiS.U.JadhavD.P.TamboliS.P.GovindwarDecolorization and detoxification of Congo red and textile industry effluent by an isolated bacterium Pseudomonas sp. SU-EBTBiodegradation2122010283296
- S.BlumelH.J.KnackmussA.StolzMolecular cloning and characterization of the gene coding for the aerobic azoreductase from Xenophilus azovorans KF46FAppl Environ Microbiol688200239483955
- A.StolzBasic and applied aspects in the microbial degradation of azo dyesAppl Microbiol Biotechnol56120016980
- A.E-L.HeshamA.M.MawadY.M.MostafaA.ShoreitStudy of enhancement and inhibition phenomena and genes relating to degradation of petroleum polycyclic aromatic hydrocarbons in isolated bacteriaMicrobiology.8352014599607
- M.MagrayA.KumarA.K.RawatS.SrivastavaP.KangueaneIdentification of Escherichia coli through analysis of 16 S rRNA and 16 S-23 S rRNA internal transcribed spacer region sequencesBioinformation6102011370371
- S.Garcia-SeguraE.BrillasCombustion of textile monoazo, diazo and triazo dyes by solar photoelectro-Fenton: decolorization, kinetics and degradation routesAppl Catal B1812016681691
- M.M.BradfordA rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye bindingAnal Biochem721976248254
- M.ZhaoP.-F.SunL-N.Du,G.WangX-M.JiaY-H.ZhaoBiodegradation of methyl red by Bacillus sp. strain UN2: decolorization capacity, metabolites characterization, and enzyme analysisEnviron Sci Pollut Res219201461366145
- G.WilkinsonStatistical estimations in enzyme kineticsBiochem J8021961324
- P.WooS.LauJ.TengH.TseK.Y.YuenThen and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratoriesClin Microbiol Infect14102008908934
- J.-S.ChangC.ChouS.-Y.ChenDecolorization of azo dyes with immobilized Pseudomonas luteolaProcess Biochem3682001757763
- J.S.ChangC.ChouY.C.LinP.J.LinJ.Y.HoT.L.HuKinetic characteristics of bacterial azo-dye decolorization by Pseudomonas luteolaWater Res3512200128412850
- M.LeelakriangsakN.T.T.HuyenS.TöweN.Van DuyD.BecherM.Heckeret al.Regulation of quinone detoxification by the thiol stress sensing DUF24/MarR-like repressor, YodB in Bacillus subtilisMol Microbiol675200811081124
- S.TöweM.LeelakriangsakK.KobayashiN.Van DuyM.HeckerP.Zuberet al.The MarR-type repressor MhqR (YkvE) regulates multiple dioxygenases/glyoxalases and an azoreductase which confer resistance to 2-methylhydroquinone and catechol in Bacillus subtilisMol Microbiol66120074054
- M.P.ShahK.A.PatelS.S.NairA.DarjiS.MaharaulMicrobial degradation of azo dye by Pseudomonas spp. MPS-2 by an application of sequential microaerophilic & aerobic processAm J Microbiol Res142013105112
- N.SupakaK.JuntongjinS.DamronglerdM.-L.DeliaP.StrehaianoMicrobial decolorization of reactive azo dyes in a sequential anaerobic–aerobic systemChem Eng J9922004169176
- S.DellerS.SollnerR.Trenker-El-ToukhyI.JelesarovG.M.GubitzP.MacherouxCharacterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilisBiochemistry4523200670837091
- M.LeelakriangsakS.BorisutCharacterization of the decolorizing activity of azo dyes by Bacillus subtilis azoreductase AzoR1Songklanakarin J Sci Technol3452012509516