Abstract
Clostridium perfringens is one of the most important globally recognised gastroenteric pathogen in humans as well as animals. The present study was aimed to know the similarities/divergence among C. perfringens type A isolates of human and animal origin using the pulsed-field gel electrophoresis (PFGE) as a molecular tool. The enterotoxic isolates obtained by screening of human diarrhoeal cases (n = 130), diarrhoeal cases of pig (n = 52) and goat (n = 50), meat samples viz., pork (n = 59) and chevon (n = 57) were characterized by standard cultural and biochemical methods followed by PCR Assays. Accordingly, a total of 11 C. perfringens type A characterized isolates (16S rRNA+, cpa+, cpb2+ and cpe+) recovered from human diarrhoeal cases (n = 3); diarrhoeal cases of pig (n = 2) and goat (n = 2); meat samples viz. pork (n = 2) and chevon (n = 2) were examined employing PFGE. The observed clustering pattern in PFGE analysis showed the relatedness between isolates from diarrhoeal goat and chevon (90–100%); diarrhoeal pig and pork (65–68%); moreover, isolates from human diarrhoeal cases were exhibiting lineage to cases from goat and pig diarrhoea as well pork and chevon by 62–68% relatedness. The outcome of the present study indicates the probable contamination of this pathogen to the human food chain through faeces from suspected food animals viz. goat and pig and their improperly cooked meat.
1 Introduction
Clostridium perfringens is one of the most important pathogen responsible for intestinal infections as well as the histotoxic diseases in humans as well as animals [Citation1]. In developed countries, C. perfringens type A food poisoning is one of the most commonly reported food-borne illness, whereas, the disease is not much explored in developing countries, including India [Citation2,Citation3].
Based on the production of four major lethal toxins i.e. alpha (α), beta (β), epsilon (ε) and iota (ι), C. perfringens is divided into five major toxinotypes/biotypes (A–E) [Citation4,Citation5]. Apart from these toxins, the pathogen also produces enterotoxin (CPE) and beta2 (β2) toxins (CPB2), which are strongly associated with food poisoning in humans and gastroenteritis in animals [Citation6]. Enterotoxigenic C. perfringens type A is also associated with cases of antibiotic associated diarrhoea (AAD) and sporadic diarrhoea [Citation7,Citation8]. Foodborne isolates of C. perfringens harbouring most frequently a chromosomal enterotoxin gene (cpe) whereas isolates from patients with AAD, sporadic diarrhoea, as well as in animal diarrhoeal cases usually have a plasmid borne cpe gene [Citation4]. The food poisoning associated with C. perfringens type A mainly occurs due to the improperly cooked and/or mishandled meat and meat products and may involve large numbers of victims [Citation9]. The source of these food poisoning outbreaks can be traced out employing appropriate epidemiological methods [Citation10]. Among various available molecular typing tools, pulsed-field gel electrophoresis (PFGE) remains one of the most important third-generation molecular typing approach for the detection of genotypic diversity of almost all bacterial species and continues to be recognized as the gold standard due to outcome spanning exceeding 90% of the bacterial genome and its standardized protocols and reagents applicable to a wide range of organisms [Citation11–Citation13]. Globally, limited studies had been carried out to assess the genetic relatedness of C. perfringens isolates from different sources [Citation14–Citation21]. But to the best of our knowledge there are no available studies regarding molecular typing of C. perfringens type A isolates through PFGE from different sources from India. With these views, the present investigation was envisaged to assess the genetic relatedness of the C. perfringens type A isolates recovered from samples of human and animal origin using PFGE as a molecular tool and thereby, to ascertain the possibility of their intersource transmission/contamination.
2 Materials and methods
2.1 Sample collection and area of the study
In the present study stool/faecal samples from human diarrhoeal cases (n = 130), diarrhoeal pig (n = 52), diarrhoeal goat (n = 50), and meat samples viz. pork (n = 59) and chevon (n = 57) were collected to know the prevalence of enterotoxigenic C. perfringens type A. The human diarrhoeal samples were collected from the National Institute of Cholera and Enteric Diseases (NICED), Kolkata, India in screw cap polypropylene tubes containing Cary-Blair transport medium by following the ethical guidelines. Animal samples were collected from organised and unorganised pig and goat farms and butcher’s herds in and around the area of humans sample source using sterile screw cap glass tubes. The pork and chevon samples were collected in sterile plastic sachet. Animal samples were also collected with the consent of the concerned authorities, whenever required. The collected samples were immediately transported to the laboratory using ice pack container and processed within 24 h for isolation and identification of C. perfringens type A using standard bacteriological and molecular procedures viz., PCR Assays.
2.2 Enterotoxigenic C. perfringens type A isolates
A total of 11 enterotoxigenic C. perfringens type A isolates (16S rRNA+, cpa+, cpb2+ and cpe+) obtained from human diarrhoeal cases (n = 3), diarrhoeal cases of pig (n = 2) and goat (n = 2) and their meat viz., pork (n = 2) and chevon (n = 2), that were closely linked to the vicinity of human diarrhoeal patients, were subjected to PFGE in order to assess the relatedness between the isolates and the possibility of cross contamination. The PFGE was performed following the PulseNet protocol [Citation22]. In brief, test strains grown on sulphite polymyxin sulphadiazine (SPS) agar (HiMedia, India) were inoculated in Robertson cooked meat medium (RCM) broth (HiMedia, India) and incubated overnight at 37°C under anaerobic condition. Then the overnight culture was suspended in Cell Suspension Buffer (CSB) (100 mM Tris:100 mM EDTA, pH 8.0) and measured the cell concentration in UV-Spectrophotometer by taking the OD value between 1.3 and 1.5 at 600 nm.
2.3 PFGE analysis
The agarose plugs were prepared by mixing equal volume of bacterial culture solution with 1% low-melting agarose (SeaKem). After solidification, the plugs were dressed following instructions with proper cutter. The bacterial cells in the agarose plugs were lysed by treatment with cell lysis buffer (50 mM Tris:50 mM EDTA, pH 8.0 + 1% Sarcosyl) and treated with proteinase K (20 mg/mL) at 54°C for 1 h with constant and vigorous agitation (150–175 rpm).
The plugs were washed twice with sterile ultrapure water (pre-heated to 50°C) under vigorous shaking in a 50°C water bath for 10–15 min each time and further washed 4 times similarly with pre-heated (50°C) TE buffer (10 mM Tris:1 mM EDTA, pH 8.0). Agarose plugs containing genomic DNA were equilibrated in TE buffer and were placed in 30 μL of 10× H buffer (0.1% BSA, 0.1% Triton X-100) [pre-incubation for digestion] for 45 min. After incubation each plug was kept overnight in 150 μL reaction mixture consisting 15 μL 10× H buffer, 15 μL 10× BSA, 3 μL SmaI enzyme (45 units) [Takara, Shuzo Co. Ltd, Japan] and 117 μL sterile triple distilled water at 37 °C.
PFGE of the SmaI digested inserts was performed by the contour clamped homogeneous electric field method on a CHEF Mapper system (Bio-Rad, California, USA) with 1% PFGE grade agarose in 0.5× TBE (40 mM Tris-HCl, pH 8.3, 45 mM boric acid, 1 mM EDTA) for 24 h using the XbaI digested DNA of Salmonella enteric serovar Braenderup (H9812) as the standard size DNA molecular marker. A mini chiller (Bio-Rad) was used to maintain the temperature of the buffer at 14°C. Run conditions (150 mA current, voltage-6.0 V/cm, angle-120°, initial switch time-10 s, final switch times-35 s, linear) were generated by the auto-algorithm mode of the CHEF Mapper PFGE system by using a 78–390 kb size range. After electrophoresis, the gel was stained in ethidium bromide (1 µg/mL) for 30 min and destained in water for 15 min twice. The DNA bands were visualized and photographed with the BioSpectrum AC Imaging System (USA). The PFGE analysis has been carried out by using BioNumerics software version 4.0 (Applied Maths, Sint-Martens-Latem, Belgium). The similarities between isolates were evaluated by using the cluster analysis with the unweighted pair group method with arithmetic mean (UPGMA) method and the Dice correlation coefficient with a position tolerance of 1.5%.
3 Results
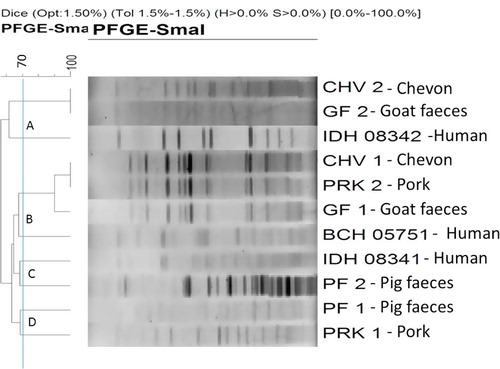
On PFGE analysis, the 11 tested isolates of C. perfringens type A (human, pig and goat diarrhoeal cases; pork and chevon) have been clustered into 4 major groups (‘A’ through ‘D’) (). The isolates from chevon (CHV2), goat faeces (GF2) and human diarrhoeal cases (IDH 08342) were belonged to the cluster ‘A’. Among them, the isolate from chevon (CHV2) and goat faeces (GF2) were grouped in a single clad and showed 100% similarity in band pattern. Further, the human isolate (IDH 08342) was linked to them with a similarity of about 62%. Isolates from four sources i.e., chevon (CHV1), pork (PRK2), goat faeces (GF1) and human diarrhoeal cases (BCH 05751) were grouped in a single cluster (cluster ‘B’); where the isolate from chevon (CHV1) and pork (PRK2) belonged to the same clad with 100% similarity; however, the isolate from goat faeces (GF1) was showing the similarity of 90% with chevon (CHV1) and pork (PRK2) isolates. Further, human isolate (BCH 05751) was found to be similar (about 68%) with these three isolates (CHV1, PRK2 and GF1, ). Further, the isolates belonging to the cluster ‘C’ [human diarrhoeal cases (IDH 08341) and pig faeces (PF2)] were found to exhibit about 68% similarity. Similarly in the cluster ‘D’, isolate from pig faeces (PF1) and pork (PRK1) was grouped with about 68% similarity. The isolates belonged to cluster ‘B’ (chevon, pork, goat faeces and human diarrhoeal cases) were linked to the cluster ‘C’ (human diarrhoeal cases and pig faeces) with a similarity range of about 65%. Similarly, the isolates from cluster ‘D’ (pig faeces and pork) were linked to isolates of cluster ‘B’ (chevon, pork, goat faeces and human diarrhoeal cases) with a similarity range of about 65%. Likewise, the isolates from human and animal origin (cluster ‘B’, ‘C’ and ‘D’) were linked to the cluster ‘A’ and expressed about 55% similarity ().
4 Discussion
On analysis of dendogram, it was observed that the isolates from chevon (CHV2), goat faeces (GF2) and human diarrhoeal cases (IDH 08342) were grouped into the cluster ‘A’. The isolates from chevon (CHV2) and goat faeces (GF2) were grouped in a single clad and showed 100% similarity in band pattern. Besides, the human isolate (IDH 08342) was linked to them with a similarity of about 62%. The belonging of the isolates from goat faeces and chevon in the same clad and band pattern similarity strongly suggests the possibility of contamination of this pathogen from food animal (goat) to its meat (chevon). This might be possible for the unhygienic dressing of carcass and its transportation. Further, the human diarrhoeal isolate (IDH 08342) showed the similarity of about 62% to the isolate from the goat faeces (GF2) and chevon (CHV2) which support the probable entry of the pathogen to human food chain through suspected goat meat (chevon). The findings of the present study were concurrent with earlier study reports [Citation18,Citation21,Citation23].
In cluster ‘B’ isolates from four sources i.e., chevon (CHV1), pork (PRK2), goat faeces (GF1) and human diarrhoeal cases (BCH 05751) were grouped and the isolates from chevon (CHV1) and pork (PRK2) were belonged to the same clad with 100% similarity; however, the isolate from goat faeces (GF1) showed similarity of 90% with chevon (CHV1) and pork (PRK2) isolates. Moreover, one human isolate (BCH 05751) was expressed about 68% similarity with these three isolates (CHV1, PRK2 and GF1). The findings showed the significant possibility for passing of this pathogen from goat to its meat. Moreover, belonging of the isolates from two meat sources i.e., chevon and pork in a same clad with 100% similarity draws attention for the possibility of cross-contamination, even these two different types of meat are separately prepared from two different type of food animals i.e., goat and pig. This could be attributed to unhygienic handling of meat during marketing chain. The genetic diversity of C. perfringens type A isolates from different sources were found to be in concurrent with the observation of genetic variation among C. perfringens isolates by using PFGE tool by others researchers [Citation21,Citation24].
Isolates from human diarrhoeal cases (IDH 08341) and pig faeces (PF2) were found to exhibit about 68% similarity in cluster ‘C’. Similar to the cluster ‘A’ and ‘B’ observations, the findings in cluster ‘C’ also expressed the strong possibility of transmission of this pathogen from pig to human or vice versa. Each isolate from pig faeces (PF1) and pork (PRK1) was grouped in cluster ‘D’ and showed about 68% similarity among them. As revealed in cluster ‘A’, the findings suggesting the probable route of contamination of this pathogen from food animal (pig) to its meat (pork). The findings of present study were concurrent with earlier study reports [Citation18,Citation20].
The analysis of PFGE findings of the isolates from human and animal sources clearly revealed their relatedness in a range of 62–68%. This indicates the probable circulation of this pathogen to the human food chain through faeces of suspected food animals viz. goat and pig and their improperly cooked meat. The present study was in accord with the earlier studies using PFGE as an epidemiological tool to track the source of infection in case of C. perfringens [Citation16,Citation19–Citation21,Citation25,Citation26].
5 Conclusions
The present study involving PFGE analysis of the tested C. perfringens type A isolates revealed the probability of contamination of meat with unhygienic slaughter and meat processing practices. It is well established that this pathogen and its enterotoxin are labile to the degree of temperature attained in the Indian traditional cuisine. The essence of the present study suggests that the adoption of standard cooking methods is indispensable to evade health hazards with this food borne zoonotic bacterial pathogen.
Acknowledgement
The authors thank the Director, Indian Veterinary Research Institute, Izatnagar, for providing necessary facilities for conducting the research work.
Competing interests
The authors declare that there are no competing interests.
Notes
Peer review under responsibility of Faculty of Veterinary Medicine, Cairo University.
References
- N.A.EzzeldeenA.M.AmmarB.ShalabyM.E.HaririrW.S.OmarRapid detection of Clostridium perfringens in seafoodAdv Environ Biol102016174181
- M.LindströmA.HeikinheimoP.LahtiH.KorkealaNovel insights into the epidemiology of Clostridium perfringens type A food poisoningFood Microbiol282011192198
- J.P.YadavS.C.DasP.DhakaD.VijayM.KumarA.K.MukhopadhyayMolecular characterization and antimicrobial resistance profile of Clostridium perfringens type A isolates from humans, animals, fish and their environmentAnaerobe472017120124
- J.LiV.AdamsT.L.BannamK.MiyamotoJ.P.GarciaF.A.UzalToxin plasmids of Clostridium perfringensMicrobiol Mol Biol Rev7722013208233
- R.O.S.SilvaF.C.F.LobatoClostridium perfringens: a review of enteric diseases in dogs, cats and wild animalsAnaerobe3320151417
- D.J.FisherK.MiyamotoB.HarrisonS.AkimotoM.R.SarkerB.A.McClaneAssociation of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin geneMol Microbiol562005747762
- A.BanaszkiewiczJ.KądzielskaA.GawrońskaH.PituchP.Obuch-WoszczatyńskiP.AlbrechtEnterotoxigenic Clostridium perfringens infection and pediatric patients with inflammatory bowel diseaseJ. Crohns Colitis82014276281
- J.C.FreedmanA.ShresthaB.A.McClaneClostridium perfringens enterotoxin: action, genetics, and translational applicationsToxins8201673
- B.M.LundT.C.Baird-ParkerG.W.GouldThe microbiological safety and quality of food2000Aspen PublishersGaithersburg, MD, USA
- S.R.PalmerEpidemiological methods in the investigation of food poisoning outbreaksLett Appl Microbiol111990109115
- E.S.NassonovaPulsed field gel electrophoresis: theory, instruments and applicationCell Tissue Biol22008557565
- R.V.GoeringPulsed field gel electrophoresis: a review of application and interpretation in the molecular epidemiology of infectious diseaseInfect Genet Evol102010866875
- R.V.GoeringP.D.FeyPulsed field gel electrophoresis of Staphylococcus epidermidisMethods Mol Biol1120145560
- S.E.MaslankaJ.G.KerrG.WilliamsJ.M.BarbareeL.A.CarsonJ.M.MillerMolecular subtyping of Clostridium perfringens by pulsed-field gel electrophoresis to facilitate food-borne-disease outbreak investigationsJ Clin Microbiol37199922092214
- B.SchalchB.SpernerH.EisgruberA.StolleMolecular methods for the analysis of Clostridium perfringens relevant to food hygieneFEMS Immunol Med Microbiol241999281286
- S.LukinmaaE.TakkunenA.SiitonenMolecular epidemiology of Clostridium perfringens related to food-borne outbreaks of disease in Finland from 1984 to 1999Appl Environ Microbiol68200237443749
- B.SchalchL.BaderH.P.SchauR.BergmannA.RometschG.MaydlMolecular typing of Clostridium perfringens from a food-borne disease outbreak in a nursing home: ribotyping versus pulsed-field gel electrophoresisJ Clin Microbiol412003892895
- A.JohanssonA.AspanE.BaggeV.BåverudB.E.EngströmK.E.JohanssonGenetic diversity of Clostridium perfringens type A isolates from animals, food poisoning outbreaks and sludgeBMC Microbiol6200647
- T.S.P.FerreiraA.M.MorenoR.R.D.AlmeidaC.R.GomesD.D.S.D.GobbiP.H.N.D.L.FilsnerMolecular typing of Clostridium perfringens isolated from swine in slaughterhouses from São Paulo State, BrazilCienc Rural42201214501456
- K.E.LeeS.I.LimS.H.ShinY.K.KwonH.Y.KimJ.Y.SongDistribution of Clostridium perfringens isolates from piglets in South KoreaJ Vet Med Sci762014745
- M.ParkJ.DeckS.L.FoleyR.NayakJ.G.SongerJ.R.SeibelDiversity of Clostridium perfringens isolates from various sources and prevalence of conjugative plasmidsAnaerobe3820162535
- CDC. http://www.cdc.gov/pulsenet/Ellipsis/PulseNetAsia_Pacific; 2009.
- M.Brzychczy-WłochM.BulandaAnalysis of genetic similarities between Clostridium perfringens isolates isolated from patients with gas gangrene and from hospital environment conducted with the use of the PFGE methodPol J Surg862014141146
- J.Y.ParkS.KimJ.Y.OhH.R.KimI.JangH.S.LeeCharacterization of Clostridium perfringens isolates obtained from 2010 to 2012 from chickens with necrotic enteritis in KoreaPoult Sci94201511581164
- P.LahtiA.HeikinheimoT.JohanssonH.KorkealaClostridium perfringens type A strains carrying a plasmid-borne enterotoxin gene (Genotype IS1151-cpe or IS1470-like-cpe) as a common cause of food poisoningJ Clin Microbiol462008371373
- A.JohanssonA.AspánM.KaldhusdalB.E.EngströmGenetic diversity and prevalence of netB in Clostridium perfringens isolated from a broiler flock affected by mild necrotic enteritisVet Microbiol14420108792