Abstract
Staphylococcus aureus is a common causative agent of bovine mastitis in dairy herds worldwide. This study was designed to assess the prevalence of mastitis in cows through screening tests and molecular characterization of Staphylococcus aureus strains. Out of 175 randomly screened cows, mastitis was detected in 50 cows by California Mastitis Test (CMT), and from those mastitic cows, 200 quarter milk samples were collected for subsequent culture and PCR based identification. The herd, cow and quarter level prevalence of mastitis was 73.3, 28.6 and 29.5% respectively, and subclinical mastitis (SCM) was the predominant type in all cases. According to bacteriology the overall prevalence of herd, cow and quarter level Staphylococcus aureus mastitis was 72.7, 74.0 and 62.0%, respectively, and the pathogen was mostly associated with clinical mastitis (CM). Cows breed, parity, daily milk yield, regular teat dipping, and dry cow therapy were significantly associated (P < 0.05) risk factors for mastitis onset. This study identifies 145 Staphylococcus aureus isolates which varied greatly with the categories of mastitis (higher in CM), udder quarter location (highest in right rear quarters), and to a lesser extent in the study areas (P < 0.05). Antimicrobial susceptibility testing revealed that 79.3% Staphylococcus aureus strains were resistant to at least one antimicrobial, 49.0% to two or more antimicrobials, and clinical isolates showed more resistance to all tested antibiotics. The highest resistance rate was found to oxytetracyclin, and no resistance to ceftriaxone and azithromycin. Seven enterotoxin gene profile were detected in the tested isolates, and mecA was found in 20.0% isolates indicating the emergence and spread of methicillin-resistant Staphylococcus aureus (MRSA). The isolates were carrying genes in combination, and were found higher in SCM cases. In this study, plasmids (>23 kb to 2.9 kb) were detected in 70.3% strains, and 54.9% plasmid bearing strains were multiple drug resistant (MDR). Thus, the high prevalence of Staphylococcus aureus mastitis is an important concern for diary industry of Bangladesh since the strains of this pathogen is becoming more resistant to commercially available antimicrobials, and this is an alarming concern for both animal and public health.
1 Introduction
Mastitis has a profound impact on dairy production, milk quality, animal health and welfare, and causes considerable economic losses to the dairy holders [Citation1]. Staphylococcus aureus is most frequent cause of mastitis in dairy animals, which is often difficult to cure and is prone to resurgence [Citation2,Citation3]. This pathogen is an increasingly recognized and most frequently isolated etiology of bovine mastitis in most countries [Citation4]. Most of the dairy animal researchers consider this organism as the true mastitis pathogens with important virulence factors [Citation3], a high level of antimicrobial resistance [Citation5], and the ability to cause chronic infections [Citation4,Citation6]. Intramammary infections (IMIs) caused by this bacterium are highly transmittable, especially during milking [Citation7]. Once established, this fearsome pathogen usually does not respond to antibiotic treatment, and in most cases treatment is associated with poor success leading to a relatively high culling rate [Citation8]. Furthermore, the treatment efficacy against this organism is usually disappointing since it causes great damages in the glandular tissues of udder, and thus, most of the antimicrobials are not able to penetrate all infected sites [Citation9,Citation10]. This bacterium also suppresses phagocytosis and cell mediated immunity, and produces an enzyme that inactivates most penicillin based treatments [Citation11]. In recent years, the emergence and spread of antimicrobials resistant Staphylococcus aureus strains, especially multidrug resistant (MDR) strains have become a major public health concern [Citation12].
Studies from Asian countries also reported Staphylococcus aureus as the chief etiologic agent of mastitis in cattle and buffalo [Citation13]. Cumbersome prevention and control of mastitis caused by this bacterium can be achieved through proper isolation, and characterization of the strains, segregation of the infected animals, dry cow therapy, treatment of clinical cases during lactation and culling program. Thus, steadfast and speedy methods for detection of Staphylococcus aureus in mastitic milk samples are crucial for the control of this disease, and economically sound udder health management [Citation14]. However, phenotypic characterization of Staphylococcus aureus is no longer beneficial in controlling mastitis caused by this organism since inter-strain variations exist in terms of virulence potential [Citation1,Citation14]. Molecular diagnostic methods like DNA-based mastitis diagnostic system have already been introduced for routine use in the dairy herds [Citation15]. Recently, PCR has become a very popular molecular technique, especially for the detection and identification of bacteria in mastitic milk by targeting their specific genes in the DNAs [Citation16,Citation17]. Therefore, the present study was designed to estimate the prevalence of bovine mastitis at herd-and cow level, and characterize the strains of Staphylococcus aureus in milk from cows having mastitis through conventional bacteriology and molecular approaches.
2 Materials and methods
2.1 Ethical approval
All the procedures of the study were performed under the approval of Bangabandhu Sheikh Mujibur Rahman Agricultural University’s Animal Experimentation Ethics Committee.
2.2 Study area
The present research work was conducted in three districts (Chittagong, Mymensingh and Gazipur) of Bangladesh during July 2015 to June 2016. The geographic position of the study area is Latitude: 20°45′–26°40′ N, Longitude: 88°05′–92°40′ E. The average annual rainfall is 3,450 mm. The day temperature ranges from 7 to 20 °C in the cool months (November to February), and in the other months it varies between 23 and 32 °C.
2.3 Study population and farm management
A total of 45 small-holding dairy farms were selected from the study areas which had previous history of mastitis, and the mean farm size was 12 (range; 5–26). In total, 175 lactating cows were randomly screened for mastitis, of which 50 were mastitis positive, and were included to this study. Most of them (74.2%) were cross-breeds (Holstein × Zebu, Sahiwal × Zebu) whilst the rest (25.8%) were local breeds (Zebu and Red Chittagong breeds). With regard to management, 14 (31.1%) of the farms were managed intensively while 31 (68.9%) farms were semi-intensive. The intensively managed cattle were kept indoors, and received concentrate feeds in addition to hay, green grass and crop residues (such as corn stalks, wheat/barley straw and other leftovers from grain threshing). On the other hand, the semi-intensively managed cattle grazed freely on pasture, but received supplementary feeds in the morning and evening when they were milked. The parity of the selected cows ranged from 1 to 5 with an average milk production 8.5 L per cow per day (range 2.0–17.0 L). The cows gave birth randomly throughout the year (no particular control breeding), were milked once daily with their calves used for stimulating milk let-down. Calves survived on residual milk after the hand milking. Control weaning was not practiced. The cows were milked manually, and the milkers did not wear gloves during the milking procedure. Pre- and post milking teat disinfection and dry cow therapy were not practiced in the study farms. Cows were housed in open shed with brick made floor, and most of the floors were wet and soiled with feces.
2.4 California mastitis test (CMT) and sample collection
CMT was used as a screening test for mastitis. It was carried out according to the procedure described by Hoque et al. [Citation18]. The CMT results were scored as 0(negative), trace, 1(weak positive), 2(distinct positive) and 3(strong positive) based on gel formation. The CMT score of 0 was considered as negative while CMT scores of 1 and 2 were considered indicators of subclinical mastitis, and 3 for clinical mastitis. Positive cows were defined as having at least one quarter with CMT score of >1. In total, 200 quarter milk (80 clinical and 120 subclinical) samples from 50 CMT positive cows were collected aseptically into the sterile plastic tubes (10–15 mL/sample). Sampling was done from all quarters of CMT positive cows, and was transported to the laboratory using ice-box.
2.5 Isolation and identification of Staphylococcus aureus
To identify the chief etiology, milk samples were collected from cows assuming that the causative organisms within a herd are similar, and also to reduce the time, labor and cost burdens. In case where only one mastitis case found in a farm, that positive cow was directly sampled. Bacteriological examination was performed within 24 h of sampling following the method described previously by [Citation3,Citation12] with some modifications. In brief, 25 mL of collected milk sample were placed into a sterile glass flask containing 225 mL of buffered peptone water (BPW, Difco, Cockeysville, MD). The solution was incubated at 37 °C in a water bath with shaking at 100 rpm for 24 h. After pre-enrichment, a 5 mL aliquot was transferred to 50 mL of trypticase soy broth (TSB, Beijing LB Technology Ltd., China) containing 7.5% NaCl. After 18–24 h incubation at 35 °C, a loopful of the culture was inoculated onto Baird-Parker agar (BPA, Beijing LB Technology Ltd., China) plates with 5.0% egg yolk and tellurite. Following incubation at 35 °C for 24 h, one or two presumptive coagulase-positive colonies per sample (black colonies surrounded by 2–5 mm clear zones) were transferred to trypticase soy agar (TSA, Beijing LB Technology Ltd., China) plates with 0.6% yeast extract for further purification. Colonies suspected of being Staphylococci were initially identified by their colony morphology and Gram staining. Catalase activity and coagulase tests were performed to distinguish catalase-negative Streptococcus spp. from catalase-positive, coagulase production by coagulase-positive Staphylococci was examined using the tube coagulation method. Then, all initially identified isolates were further confirmed as Staphylococcus by multiplex PCR detection (at least two times confirmation) using genus specific oligoneucleotide primers as previously described by Wang et al. [Citation12]. Colonies were confirmed as Staphylococcus aureus by PCR detection of the thermonuclease gene (nuc; Staphylococcus aureus specific gene). Finally, all isolates were stored in brain heart infusion broth with 15.0% glycerol at −80 °C until further use.
2.6 Antimicrobial susceptibility testing
The susceptibility of the isolates to various commonly used antimicrobials was performed by using both disk diffusion and agar dilution methods according to the guidelines of the Clinical Laboratory Standards Institute [Citation19]. The disk diffusion test was performed following the procedure described by [Citation10]. The agar dilution method was used to measure the MICs of penicillin, erythromicin, oxytetracycline, trimethoprim/sulfamethoxazole, ciprofloxacin, gentamicin, amoxicillin, oxacillin, ceftriaxone and azithromycin [Citation10,Citation12]. The breakpoints of CSLI for the tested antimicrobials (for both disk diffusion and agar dilution) were used to determine the susceptibility profiles. All antimicrobial susceptibility testing assays were repeated at least 3 times. E. coli ATCC 25,922 and Staphylococcus aureus ATCC 29,213 were included as quality control strains in each run [Citation12].
2.7 DNA extraction and purification
The chromosomal DNA of Staphylococcus aureus was extracted by boiling method by following the methods of Aldous et al. [Citation20] with some modifications. Well isolated single colony from the Baird Parker agar was sub-cultured onto nutrient agar (NA) plate, and kept overnight incubation at 37 °C, and finally one pure colony from NA plate was transferred to 5 mL nutrient broth and incubated at 37 °C with aeration using shaker machine set at 120 rpm. One milliliter (1.0 mL) culture was taken in an eppendorf tube (1.5 mL), centrifuged at 13,000 rpm for 10 min, and cell pellets were collected. The cell pellets were then washed with distilled water by re-centrifugation. Then 200µL PCR water was mixed and dissolved by hand shaking. After that, each eppendorf was kept at 100 °C boiling temperature for 10 min followed by 10 min cold shock in ice. The tubes were again centrifuged at 10,000 rpm for 10 min, and the supernatant (100-150µL) was collected into a fresh eppendorf tube. DNA concentration and purity were evaluated by optical density using a NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies, Rockland, DE, USA) at different wavelengths [Citation21], and stored at −80 °C until further used for PCR.
2.8 Multiplex PCR conditions
The prevalence rates of five Staphylococcus aureus enterotoxin genes (sea, seb, sec, sed, and see), the toxic shock syndrome toxin 1 gene (tsst-1), exfoliative toxin genes (eta and etb), panton-valentine leukocidin gene (pvl) and methicillin resistant gene (mecA) were determined by multiplex PCR. The multiplex PCR was done in two separate runs to prevent mis-interpretation of closely related band lengths with primers specific for the respective genes () from the previously published sequences [Citation12,Citation22,Citation23]. Two sets of primer mixes were prepared according to the master mixes of components from the Taq DNA Polymerase kit (Promega Corp., Madison, Wisconsin, USA), with slight modifications to the given instructions. Of the 2 multiplex PCR sets, Set A was designed to test for nuc, sea, seb, sec, sed, and see, and contained 1.5 µL of each of the nuc, sea, seb, sec, sed, and see primer pairs. The PCR mixture consisted of 1.5 mM MgCl2, 10 mMTris-HCl (pH 9.0), 50 mMKCl, 0.1% Triton®X-100, 200 mM (each) deoxynucleotide triphosphate, 0.2 mM of the respective primers, and 0.625 U Taq polymerase (all the reagents from Promega Corp., Madison, Wisconsin, USA). The amplification was performed with an automated thermolcycler T-1 (Biometra). Multiplex PCR Set B constituted of the same ingredients as in set A, except for the MgCl2 concentration (2.0 mM) and the mecA, pvl, tsst 1, eta, and etb primer pairs. Three positive control containing Staphylococcus aureus reference strains (ATCC 29213; sea, seb, sec, sed, see and tsst positive, ATCC 25923; pvl positive, and ATCC 43300; mecA positive) were used in each PCR run, and however, for certain Staphylococcus aureus toxins (eta and etb) no positive control was used in PCR reactions (just water). The PCR cycles consisted of pre-heating at 95 °C for 10 min, denaturation at 94 °C for 5 min, annealing at 55 °C for 0.5 min, and extension at 72 °C for 1.5 min. The amplification was performed for 35 cycles with a final extension step at 72 °C for 3.5 min. The specificity of this PCR was evaluated with the reference strain of Staphylococcus aureus ATCC 25,923 and with all bacteria (Streptococcus spp., Micrococcus spp., coagulase negative Staphylococcus, E. coli) strains previously isolated from the milk samples [Citation12,Citation22,Citation23]. The PCR products were analyzed by electrophoresis in a 1.5% agarose gel containing 0.5 mg of ethidium bromide per mL, visualized and photographed with Image Master VDS (Pharmacia Biotech). The sizes of the amplification products were estimated by comparison with a 100 bp DNA step ladder (Promega Corp., Madison, Wisconsin, USA).
2.9 Plasmid extraction from Staphylococcus aureus isolates
Isolation of plasmid DNA in Staphylococcus aureus isolates was done according to the method previously reported by different researchers [Citation13,Citation24]. Briefly, a single colony of pure Staphylococcus aureus was inoculated into 5 mL of Luria-Bertani (LB) (Oxoid, Wesel, Germany) broth and incubated in orbital shaking incubator (Labnet 211DS, USA) (200 rpm) at 37 °C for 16 to 18 h, and then centrifuged at 4800 rpm for 5 min, and the resulting cell pellets were resuspended in 300μL, TENS buffer (Tris-EDTA-NaOH/SDS). Then, the solution was mixed for 2–3 s until the mixture became sticky. Then the samples were incubated in ice for 10 min to prevent the degradation of chromosomal DNA. Thereafter, 150 μL 3 M sodium acetate (Sigma-Aldrich, USA, pH = 5.2), was added and vortexed 2–5 s to mix completely. The mixture was spun again at 13,200 rpm for 10 min to pellet cell debris and chromosomal DNA. The supernatant was transferred into a fresh microtube and mixed with 1 mL of 95% EtOH (Ethanol) which has been pre-cooled to −20 °C and further spun for 2 min to pellet plasmid DNA and RNA. The supernatant was also discarded, and the pellet rinsed twice with 500 μL of 70% EtOH and dried at room temperature. For the subsequent steps, the isolated plasmid DNA was resuspended in 200μL of TE (Tris-EDTA) buffer; at pH = 8 and 200 ng/μLRNAse were also added. Plasmids were separated by electrophoresis in 1.5% agarose gel containing 0.5 mg of ethidium bromide per mL (all the reagents from Promega Corp., Madison, Wisconsin, USA) at a voltage of 4.5 V/cm; buffer: 1 x TAE (Tris-Acetate-EDTA); time: 3 h, and thereafter observed under UV light to visualize the bands properly. The image was recorded and analyzed using Image Master VDS (Pharmacia Biotech).
2.10 Statistical analyses
The data generated from this experiment were entered in Microsoft Excel (2010) worksheet, organized and processed for further descriptive analyses. The Cochran-Mantel-Haenszel χ2 test was performed using statistical packages for social sciences (SPSS), version 16.5 (IBM SPSS statistics for windows, Chicago, IL, USA) to compare Staphylococcus aureus intramammary infection (IMI) between the studied dairy farms and areas, and also to compare the cultural results in PCR positive and negative samples. For the test, P < 0.05 was considered statistically significant.
3 Results
3.1 Prevalence of mastitis, isolation of Staphylococcus aureus, and associated risk factors
The findings of the CMT and clinical examination confirmed overall 73.3% prevalence of bovine mastitis in the study areas. However, overall cow level and quarter level mastitis were 28.6 and 29.5% respectively, on the basis of CMT and physical scoring, of which 31.1, 38.0 and 40.0% were clinical mastitis (CM) and 42.2, 62.0 and 60.0% were subclinical mastitis (SCM), respectively (). Bacteriology confirmed overall 72.7% (24/33) prevalence of herd level mastitis caused by Staphylococcus aureus in the study areas, of which clinical and subclinical mastitis were 62.5 and 37.5%, respectively (). The study identified several potential factors on the occurrence of Staphylococcus aureus mastitis, and among those factors, breed, parity, per day milk yield, regular teat dipping and dry cow housing were found to be significantly (P < 0.05) associated with mastitis prevalence. On the other hand, body condition scores (BCS) and herd size did not have significant effect (P > 0.05) on the occurrence of mastitis (). However, the overall prevalence of cow- and quarter-level Staphylococcus aureus mastitis was 74.0 and 62.0%, respectively. In both cases, the pathogen was predominantly associated with clinical mastitis rather than subclinical mastitis.
Table 1 Herd and cow-level prevalence of mastitis (clinical and subclinical) in some selected areas of Bangladesh.
Table 2 Herd and cow-level prevalence of Staphylococcus aureus mastitis (clinical and subclinical) in some selected areas of Bangladesh.
Table 3 Odds ratios (OR) and confidence intervals (CI) of factors having significant effect on Staphylococcus aureus mastitis in dairy cows (Logistic regression model).
A total of 145 Staphylococcus aureus isolates were detected (1–2 isolates per sample), of which 85 isolates were from SCM and 60 from CM samples. Our present findings revealed that the number of Staphylococcus aureus isolates varied greatly with the categories of mastitis, and to a lesser extent in the study areas (). The number of isolates also varied according udder quarter location, and remained highest in right rear (RR; 48.0% SCM and 51.0% CM) quarters, followed by left rear (LR; 41.0% SCM and 48.0% CM) quarters, left front (LF; 28.0% SCM and 33.0% CM) quarters, right front (RF; 29.0% SCM and 22.0% CM) quarters, respectively (). At farm level, Staphylococcus aureus was isolated from 73.3% (33/45) of the farms affected with mastitis. Amplification with genus specific PCR successfully confirmed the isolates as Staphylococcus aureus by amplification of DNA fragments (nuc; 279 bp) ().
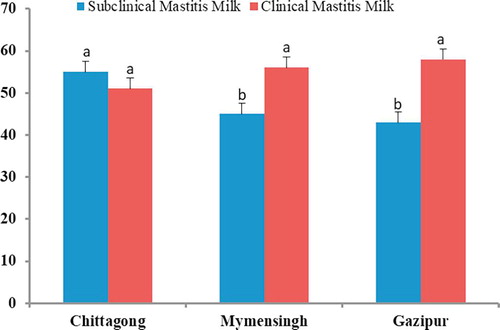
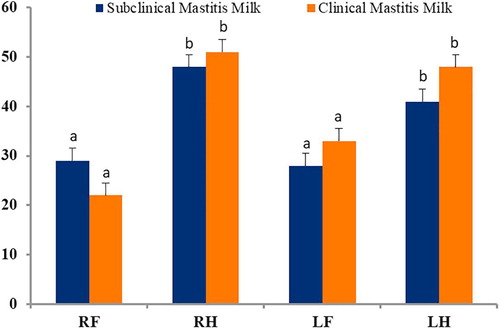
Table 4 Primer sequences, anticipated product size, and sets of multiplex PCR used in this study for identification of Staphylococcus aureus genes.
3.2 Antimicrobial susceptibility profile
Antibiotic susceptibility pattern of the analyzed Staphylococcus aureus isolates is shown in . The overall resistance rates were higher with oxytetracyclin (74.5%), followed by oxacillin (55.9%), ciprofloxacin (49.6%), amoxicillin (42.0%), trimethoprim/sulfamethoxazole (30.0%), and to a less extent to gentamicin (17.9%), penicillin (11.0%) and erythromycin (8.2%). However, none of the tested isolates were found resistant to ceftriaxone and azithromycin. Out of 145 isolates, 79.3% were resistant to at least one antimicrobial, while 49.0% to three or more antimicrobials (). Antimicrobial susceptibility pattern also varied according to the category of mastitis milk samples, 5.9 and 15.0% isolates respectively from SCM and CM cases were resistant to one antimicrobial, 20.0% and 21.7%, respectively to two antimicrobials, 12.9% and 30.0%, respectively to three antimicrobials, 23.5% and 25.0%, respectively to four antimicrobials, and 7.0% and 0.0%, respectively to five antimicrobials. Isolates from CM samples showed higher resistance to at least one antimicrobial (91.7%) than isolates recovered from SCM samples (70.6%) ().
Table 5 The overall antimicrobial susceptibility patterns of the Staphylococcus aureus isolates (n = 145) from bovine mastitis (subclinical and clinical mastitis) milk samples.
Table 6 Multidrug resistance (MDR) properties observed among 145 Staphylococcus aureus isolates in bovine mastitis (subclinical and clinical mastitis) milk samples.
3.3 PCR detection of mecA and toxin genes
Using previously published primers (), we analyzed the presence of mecA and other enterotoxin genes in Staphylococcus aureus isolates. The lists the toxin gene profile of the isolates. In our current investigation, seven different toxin gene profile in various combinations were found in the DNA samples extracted from pure isolates (single colony). Among the isolates examined in this study, the overall detection rate of mecA was 20.0% (29 out of 145), indicating the high prevalence of methicillin resistant (MRSA) strains in Staphylococcus aureus isolates derived from bovine mastitis in Bangladesh. Out of 145 tested isolates, 101 (69.7%) were positive for one or more toxin genes, and six toxin genes (pvl, sea, seb, sec, sed and see) were detected in these isolates. The three most predominant toxin genes were pvl (24.8%), sea (17.9%) and seb (11.0%), followed by sec (9.7%), see (9.7%) and sed (6.2%). In this study, 20.7% isolates were carrying two genes and the genotype sec-pvl (9.0%; 9/145) was the most common genotype. Independent origin of the milk samples exhibited that 3.4% (5/145) isolates were carrying three (sea-seb-pvl) genes and genotypes encoding four enteroxins (seb-sec-sed-pvl) were detected only in one (0.7%) isolate. Highest numbers of enterotoxin producing genes were detected in subclinical isolates (85.9%), and their existence was as either single gene or multiple genes. However, none of the 145 isolates showed the presence of the eta, etb, and tsst 1 genes. Furthermore, variation in the distribution of Staphylococcus aureus strains harboring antimicrobial resistant (AMR) genes was recorded; however, the frequency of AMR genes did not vary significantly among strains of subclinical and clinical milk samples ().
Table 7 mecA and other toxin gene profile found in Staphylococcus aureus isolates in bovine mastitis (subclinical and clinical) milk samples.
3.4 Plasmid profile analysis
Among 145 Staphylococcus aureus isolates, plasmids were detected in 70.3% (102/145) strains. The molecular weight of plasmids varied from >23 kb to 2.9 kb. Most of the isolates showed only single plasmid band with size of 18.4 kb (67.6%) while the rest of the strains had 2 to 4 plasmids ranging from >23 kb to 2.9 kb (32.4%). However, the most common plasmid of 18.4 kb was detected in all strains. Antimicrobial susceptibility tests showed that 54.9% (56/102) of these plasmid bearing strains were multiple drug resistant (MDR) and the rest 45.0% plasmids were susceptible to all the tested antimicrobials. In contrast, 29.7% (43/145) isolates had no plasmid DNA and among these isolates, 15 were resistant to at least three or more antimicrobials. Thus, the numbers of plasmids found in a particular isolate would not necessarily indicate the level of MDR properties of the isolate.
4 Discussion
Globally, mastitis is the most fearsome infectious disease affecting dairy cattle and remains as a constant challenge in the dairy industry. Successful management, prevention and treatment of bovine mastitis are great inevitable task for the dairy holders. Staphylococcus aureus mastitis outcomes are highly variable and depend, in part, on strain dependent features. However, reports on Staphylococcus aureus contamination of bovine milk are scarce in Bangladesh and this is the first ever comprehensive investigation on molecular characterization of Staphylococcus aureus strains in milk from bovine mastitis.
Our current investigation is one of the few bovine mastitis (both SCM and CM) studies in Bangladesh in which prevalence of overall mastitis at both cow and farm level and their associated risk factors has been studied, and as well the most predominant etiology has been identified and characterized. The study revealed that 73.3% of the farms observed had at least a cow suffering from mastitis. However, our present findings of the prevalence of farm-level bovine mastitis could not be compared with other mastitis studies of Bangladesh due to lack of similarly designed researches. The overall cow-level prevalence of mastitis was 28.6%, of which majority of the cows (62.0%) were suffering from subclinical mastitis (SCM), and rest 38.0% of cows were affected with clinical mastitis (CM). The prevalence of quarter level mastitis in this study was 29.5% (60.0%; SCM and 40.0%; CM). Our current results are within the range of cow-level mastitis prevalence (8.0–64.0%) reported by most recently published studies in the country [Citation18,Citation25,Citation26]. Several earlier findings and our observation are higher relative to the available reports from other Asian countries whose dairy management is more or less similar to ours, i.e., 63.8% SCM in Thailand [Citation27], 18.2% CM and 33.7% SCM in Pakistan [Citation28], 30.6–33.7% SCM [Citation29] and 16.0% CM [Citation30] in India. This entails how serious the problem is, in the dairy sector of the continent that warrant due attention.
The study found Staphylococcus aureus as chief etiology of bovine mastitis and the pathogen was isolated in 72.7% of the examined farms. In this study, the farm level prevalence of SCM and CM associated with Staphylococcus aureus was 62.5 and 37.5%, respectively. The individual cow-level and quarter-level prevalence mastitis were 74.0 and 62.0%, respectively and this bacterium was predominantly associated CM. These results are strongly supported by the recent findings of [Citation31], who also reported this pathogen as the leading causative agent for bovine mastitis in Ethiopia. Despite our current study, many of the previously published studies have also reported Staphylococcus aureus as the main etiological agent of bovine mastitis in different countries [Citation32]. However, most of the previous studies were based on phenotypic identification, which may misidentify and underestimate the true prevalence of Staphylococci mastitis. The prevalence of bovine SCM caused by Staphylococcus aureus was reported 21.2 and 31.0%, respectively by Nazneen et al. [Citation13] in different areas of Bangladesh. In another study [Citation33] reported that about 95.0% of CM in dairy cattle are caused major pathogens and Staphylococcus aureus is one of the these major pathogens [Citation34]. Previously working with bovine mastitis, [Citation35] obtained Staphylococcus aureus in 72.5 and 28.3% milk samples in Poland and Turkey, respectively. This variability between different reports could be attributed to the differences in farm management practices or to differences in study methods and instruments employed by different researchers. The high prevalence of Staphylococcus aureus in our present investigation might be associated with hygienic and management factors (like breeds, farm size, absence of teat dipping before and after milking, lack of diagnosing the subclinical and chronic forms of mastitis, absence of dry cow therapy, no diagnostic facilities, and practice of hand milking) in the studied dairy farms. Staphylococcus aureus and other contagious microorganisms are usually found on the udder or teat surface of infected cows and are the primary source of infection between uninfected and infected udder quarters, usually during milking [Citation31].
In this study, quarter wise analysis of both SCM and CM revealed that the rear quarters were more susceptible to Staphylococcus aureus mastitis. The right rear (RR) quarters were most susceptible (RR; 48.0% SCM and 51.0% CM) to infections, and in most of the cases, the CM had higher number of Staphylococcus aureus isolates than the SCM. Variation in quarter-wise and animal-wise incidence of intramammary infections (IMIs) was also reported in several earlier studies [Citation18], and maximum numbers of animals (74.3%) were having one quarter infection [Citation36]. In a previous study [Citation37] reported that 76.7% of the examined farms suffered from Staphylococcus aureus infection, and the overall prevalence of this pathogen was 12.2% in quarter milk samples. In recent years, the emergence of multiple antimicrobial resistant strains of Staphylococcus aureus, particularly MDR strains leading to either SCM or CM has become a major public health concern. The result of our current investigation revealed that most of the Staphylococcus aureus strains were highly resistant to oxytetracyclin, oxacillin, ciprofloxacin, amoxicillin, trimethoprim/sulfamethoxazole, and to a less extent to gentamicin, penicillin and erythromycin. Among the tested isolates, only two (1.4%) were sensitive to all tested antimicrobials, while 49.0% isolates showed multidrug resistance (resistant to at least three antimicrobials). These results coincide with the findings of [Citation12,Citation13] who also reported similar type of antimicrobial resistance against Staphylococcus aureus isolates. Another study from China reported 77.3% of Staphylococcus aureus isolates tested were resistant to antimicrobials [Citation36], while studies in Denmark, Brazil and Argentina reported figures of 75.0, 55.1 and 40.0% antimicrobial resistance, respectively [Citation38,Citation39]. Resistance to oxytetracycline, oxacillin, ciprofloxacin, amoxicillin, tirmethoprim/sulfamethoxazole, gentamicin, penicillin, and erythromicin (74.5, 55.9, 49.6%, 42.0, 30.0, 17.9%, 11.0 and 8.2%, respectively) was common among the isolates of both SCM and CM samples. Antimicrobial resistance of Staphylococcus aureus isolates of our current study closely proximate with the findings of Wang et al. [Citation12] and Mehrotra et al. [Citation23], and slightly higher than the findings of [Citation13]. Appearance of resistance against a particular antibiotic in a specific region may be due to its frequent and long-term use [Citation40], and isolates from different samples and even between herds in the same farm [Citation41].
Methicillin-resistant Staphylococcus aureus (MRSA) is a growing concern worldwide and the mecA-positive Staphylococcus aureus can be isolated from bovine mastitis [Citation42]. Multiplex PCR using DNA extracted from pure single colony revealed that 20.0% of the MRSA isolates were found carrying mecA genes agreeing with the findings of [Citation43], who reported 17.5% prevalence of mecA genes in bovine mastitis. Presence of mecA gene is generally recognized as the most reliable method for detection of methicillin resistance and mecA-positive Staphylococcal strains are considered to be resistant to most of the antimicrobials. In regard to the genes encoding enterotoxins, our present findings showed that 69.7% isolated strains (from pure colonies) were positive for at least one enterotoxin gene, with one strain even presenting a combination of six genes (pvl, sea, seb, sec, sed and see). A previous study, [Citation43] reported that 67.8% Staphylococcus aureus isolates were positive for the presence of genes coding for one or more enterotoxin with a frequency very close to our present findings. Although, the gene pvl alone was predominant in 24.8% of the isolates, but it was found in combination with other genes (sea, seb, sec and sed). The frequency of pvl gene either alone or in combination in our present investigation is much lower than the results reported before [Citation44], which reported that over 50.0% of bovine Staphylococcus aureus isolates were carrying pvl genes. Ironically, none of the genes encoding the classical toxins, eta, etb and tsst-1 genes was found in our present investigation and this result is in agreement with several previous studies [Citation45].
In addition, our present findings revealed that majority (70.3%) of the Staphylococcus aureus isolates were carrying one or more plasmids, and about 54.9% of these plasmid bearing isolates were resistant to multiple drugs. This finding is supported by the results of [Citation13] who reported that 58.3% of Staphylococcus aureus isolates carrying plasmids showed MDR properties. Plasmid profiling of Staphylococcus aureus strains could be an efficient method to characterize the strains, and their interaction with host cellular immune defenses in bovine mastitis [Citation24,Citation46]. Although, the subclinical isolates had higher numbers of enterotoxin producing genes than the clinical isolates, however, their antimicrobial susceptibility pattern did not differ significantly. Thus, it can be postulated that resistance properties of Staphylococcus aureus isolates are not always dependent on the presence or absence of enterotoxins and plasmids.
5 Conclusions
The present investigation has revealed that mastitis, particularly subclinical type, is a widely prevalent disease in the dairy farms of Bangladesh both at herd- and cow-level. Although not studied well, lack of routine mastitis preventive and control measures by the dairy holders, and predominating risk factors (inadequate farm management and unhygienic milking) have been noted as the main reasons for high prevalence of mastitis in the study areas. The present study explored that Staphylococcus aureus is an important cause of mastitis, which warns the higher public health risk due to consumption of raw milk and its products. The findings of the current study also exposed that a broad distribution of identical or closely related enterotoxin producing or methicillin-resistant Staphylococcus aureus strains contribute to bovine mastitis problems in dairy cows of Bangladesh. There is a need to improve the knowledge level of farmers towards mastitis management. Effort is needed to strengthen the molecular surveillance of Staphylococcus aureus associated with bovine mastitis. Therefore, careful monitoring for the resistance status is an utmost need since the transmission of this pathogen is dynamic and involves human, animals, and likely the farm production environment.
Acknowledgements
The authors of this manuscript are grateful to the research management committee (RMC) of the Bangabandhu Sheikh Mujibur Rahman Agricultural University, Gazipur 1706, and University Grants Commission (UGC), Bangladesh. We are highly thankful to the scientists of the Centre for Food and Waterborne Diseases (CFWD), International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) for their kind support to carry out the molecular works. The cooperation and support by the dairy holders during data collection and examination of animals are highly acknowledged.
Funding Source
This research work was jointly funded by the research management committee (RMC) of the Bangabandhu Sheikh Mujibur Rahman Agricultural University, Gazipur 1706, and University Grants Commission (UGC), Bangladesh (Grant No. RMC/UGC/BSMRAU/2014-15/sec.-07/Sl. No. 32).
Competing interests
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Notes
Peer review under responsibility of Faculty of Veterinary Medicine, Cairo University.
References
- C.Le MaréchalN.SeyffertJ.JardinD.HernandezG.JanmL.RaultMolecular basis of virulence in Staphylococcus aureus mastitisPLoS ONE62011e27354 10.1371/journal.pone.0027354
- K.KatsudaE.HataH.KobayashiM.KohmotoK.KawashimaH.TsunemitsuMolecular typing of Staphylococcus aureus isolated from bovine mastitic milk on the basis of toxin genes and coagulase gene polymorphismsVet Microbiol1052005301305
- S.HosseinzadehD.H.SaeiStaphylococcal species associated with bovine mastitis in the North West of Iran: Emerging of coagulase-negative StaphylococciInt J Vet Sci Med220142734
- B.E.GillespieS.I.HeadrickS.BoonyayatraS.P.OliverPrevalence and persistence of coagulase-negative Staphylococcus species in three dairy research herdsVet Microb13420096572
- Y.FreyJ.P.RodriguezA.ThomannS.SchwendenerP.VincentGenetic characterization of antimicrobial resistance in coagulase-negative Staphylococci from bovine mastitis milkJ Dairy Sci96201322472257
- D.CervinkovaH.VlkovaI.BorodacovaJ.MakovcovaV.BabakA.LorencovaPrevalence of mastitis pathogens in milk from clinically healthy cowsVet Med582013567575
- C.SterM.AllardS.BoulangerM.L.BouletJ.MulhbacherD.A.LafontaineExperimental treatment of Staphylococcus aureus bovine intramammary infection using a guanine riboswitch ligand analogJ Dairy Sci96201310001008
- R.S.HendriksenD.J.MeviusA.SchroeterC.TealeD.MeunierP.ButayePrevalence of antimicrobial resistance among bacterial pathogens isolated from cattle in different European countries: 2002–2004Acta Vet Scand50200828
- L.GülerÜ.OkK.GündüzY.GülcüH.H.HadimliAntimicrobial susceptibility and coagulase gene typing of Staphylococcus aureus isolated from bovine clinical mastitis cases in TurkeyJ Dairy Sci88201031493154
- W.PuY.SuJ.LiC.LiZ.YangH.DengHigh Incidence of Oxacillin-Susceptible mecA-Positive Staphylococcus aureus (OS-MRSA) Associated with bovine mastitis in ChinaPLoS ONE92014e88134
- A.P.De OliveiraJ.L.WattsS.A.SalmonF.M.AarestrupAntimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Europe and United StatesJ Dairy Sci832000855862
- X.WangJ.MengJ.ZhangT.ZhouY.ZhangB.YangCharacterization of Staphylococcus aureus isolated from powdered infant formula milk and infant rice cereal in ChinaInt. J Food Microb1532012142147
- N.I.NazneenZ.FarzanaA.M.M.A.ChowdhuryA.MannanK.M.KamaruddinA.M.A.M.Z.SiddikiCharacterization of bovine subclinical mastitis caused by Staphylococcus aureus in southern Bangladesh by bacteriological and molecular approachesAsian J Biol Sci72014112
- R.PillaG.G.M.SnelM.MalvisiR.PiccininiDuplex real-time PCR assay for rapid identification of Staphylococcus aureus isolates from dairy cow milkJ Dairy Res802013223226
- M.T.KoskinenJ.HolopainenS.PyöräläP.BredbackaA.PitkäläH.W.BarkemaAnalytical specificity and sensitivity of a real-time polymerase chain reaction assay for identification of bovine mastitis pathogensJ Dairy Sci922009952959
- S.TaponenL.SalmikiviH.SimojokiT.KoskinenS.PyöräläReal-time polymerase chain reaction-based identification of bacteria in milk samples from bovine clinical mastitis with no growth in conventional culturingJ Dairy Sci92200926102617
- M.S.ElsayedA.M.El-BagouryM.A.DawoudPhenotypic and genotypic detection of virulence factors of Staphylococcus aureus isolated from clinical and subclinical mastitis in cattle and water buffaloes from different farms of Sadat City in EgyptVet World8201510511058
- M.N.HoqueZ.C.DasA.K.TalukderM.S.AlamA.N.M.A.RahmanDifferent screening tests and milk somatic cell count for the prevalence of subclinical bovine mastitis in BangladeshTrop Anim Health Prod4720157986
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing 2006, Sixteenth International Ed. Document M100–S16, Pennsylvania, USA.
- W.K.AldousJ.I.PounderJ.L.CloudG.L.WoodsComparison of six methods of extracting Mycobacterium tuberculosis DNA from processed sputum for testing by quantitative real-time PCRJ Clin Microb43200524712473
- G.OikonomouM.L.BicalhoE.MeiraR.E.RossiC.FoditschV.S.MachadoMicrobiota of cow’s milk; distinguishes healthy, sub-clinically and clinically diseased quartersPLoS ONE92014e85904 10.1371/journal.pone.0085904
- F.PelesM.WagnerL.VargaI.HeinP.RieckK.GutserCharacterization of Staphylococcus aureus strains isolated from bovine milk in HungaryInt J Food Microbiol1182007186193
- M.MehrotraG.WangW.M.JohnsonMultiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistanceJ Clin Microb38200010321035
- U.M.TumlamD.R.KalorayM.P.NandePlasmid profile and antimicrobial resistance pattern of coagulase negative Staphylococci (CNS) bacteria isolated from bovine subclinical mastitisAnim Sci Pap Rep720139095
- Shamsuddin M, Bhattacharjee J, Goodgear WJ, Momont H, Frank G, Talukdar AK et al. Community-based productivity veterinary service for smallholder dairy farmers in Bangladesh. In Odongo NE, Garcia M, Viljoen GJ, editors. Sustainable improvement of animal production and health, FAO, Rome, Italy; 2010. p. 247–53.
- A.K.JhaM.N.HoqueM.M.KamalM.M.RahmanM.M.U.BhuiyanM.ShamsuddinPrevalence of mastitis and efficacy of different treatment regimens on clinical mastitis in cowsSAARC J Agric820107989
- C.JarassaengS.AiumlamaiC.WachirapakornM.TchakumphuJ.P.T.M.NoordhuizenA.C.BeynenRisk factors of sub-clinical mastitis in small-holder dairy cows in KhonKaen ProvinceThai J Vet Med422012143151
- S.HameedM.ArshadM.AshrafM.AvaisM.A.ShahidCross-sectional epidemiological studies on mastitis cattle and buffaloes of Tehsil Burewala, PakistanJ Anim Plant Sci222012371376
- A.ElangoK.A.DoraisamyG.RajarajanG.KumaresanBacteriology of sub-clinical mastitis and anti-biogram of isolates recovered from cross-bred cowsIndian J Anim Res442010280284
- Y.C.BangarM.R.VermaA.K.DohareMukherjee R.ReenaMeta-analysis of prevalence of clinical mastitis in crossbred cows in India (1995–2014)J Anim Res62016933938
- R.AbebeH.HatiyaM.AberaB.MegersaK.AsmareBovine mastitis: prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South EthiopiaBMC Vet Res122016270 10.1186/s12917-016-0905-3
- Food and Agriculture Organization (FAO). Impact of mastitis in small scale dairy production systems. Animal Production and Health Working Paper. No. 13. Rome; 2014.
- B.A.TenhagenG.KoSterJ.WallmannW.HeuwieserPrevalence of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Brandenburg, GermanyJ Dairy Sci89200625422551
- S.PiepersL.DeMeulemeesterA.deKruifG.OpsomerH.W.BarkemaS.DeVliegherPrevalence and distribution of mastitis pathogens in subclinically infected dairy cows in Flanders, BelgiumJ Dairy Res742007478483
- S.KirkanE.O.GoksoyO.KayaIdentification and antimicrobial susceptibility of Staphylococcus aureus and coagulase negative Staphylococci from bovine mastitis in the Aydin Region of TurkeyTurk J Vet Anim Sci292005791796
- N.SudhanA.R.SinghM.SinghJ.S.SoodanStudies on prevalence, etiology, and diagnosis of sub clinical mastitis among cross breed cowsIndian J Anim Res392005127130
- J.LiH.ZhouL.YuanT.HeS.HuPrevalence, genetic diversity, and antimicrobial susceptibility profiles of Staphylococcus aureus isolated from bovine mastitis in Zhejiang Province, ChinaJ Zhejiang Univ Sci102009753760
- H.W.BarkemaY.H.SchukkenR.N.ZadoksThe role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitisJ Dairy Sci89200618771895
- T.SchmidtIn vitro antimicrobial susceptibility of Staphylococcus aureus strains from dairy herds in KwaZulu-NatalJ South Afr Vet Assoc8220117679
- R.KumarB.R.YadavR.S.SinghGenetic determinants of antibiotic resistance in Staphylococcus aureus isolates from milk of mastitic crossbred cattleCur Microb602010379386
- V.L.M.RallE.S.MirandaI.G.CastilhoC.H.CamargoDiversity of Staphylococcus species and prevalence of enterotoxin genes isolated from milk of healthy cows and cows with subclinical mastitisJ Dairy Sci972013829837
- M.J.PajicZ.B.RasicB.M.VelebitS.F.BobosM.M.MihajlovicM.Z.RadinovicThe prevalence of methicillin resistance and Panton-Valentine leukocidin synthesis genes in Staphylococcus aureus isolates of bovine and human originVet Arch832014205214
- H.TurutogluS.ErcelikD.OzturkAntibiotic resistance of Staphylococcus aureus and coagulase-negative Staphylococci isolated from bovine mastitisBull Vet Inst Pulawy5020064145
- A.ZecconiL.CesarisE.LiandrisV.DapraR.PiccininiRole of several Staphylococcus aureus virulence factors on the inflammatory response in bovine mammary glandMicrob Pathogenesis402006177183
- N.ÜnalO.D.ÇinarDetection of Stapylococcal enterotoxin, methicillin-resistant and Panton-Valentine leukocidin genes in coagulase-negative Staphylococci isolated from cows and ewes with subclinical mastitisTrop Anim Health Prod442012369375
- E.ArslanA.ÇelebiL.AçikU.S.UçanCharacterization of coagulase positive Staphylococcus species isolated from bovine mastitis using protein and plasmid patternsTrop Anim Health Prod332009493500
