Abstract
Bovine norovirus (BNoV) has emerged as a viral pathogen that causes a gastrointestinal illness and diarrhea in cattle. Despite its worldwide distribution, very little information is known about BNoV in Africa. In this study, BNoV was detected in 27.6% (8/29) of tested fecal materials, collected from sporadic cases of diarrheic calves, using the reverse transcription-polymerase chain reaction (RT-PCR) and primers that target RNA dependent RNA polymerase gene. Additionally, one primer pair was designed to flank the BNoV-VP2 (small capsid protein) gene for molecular analysis. Study VP2 sequences were phylogenetically-related to BNoV-GIII.2 (Newbury2-like) genotype, which is highly prevalent all over the world. However, they were separated within the cluster and one strain (41FR) grouped with recombinant GIII.P1/GIII.2 strains. Compared to reference VP2 sequences, 14 amino acid substitution mutations were found to be unique to our strains. The study confirms that BNoV is currently circulating among diarrheic calves of Egypt and also characterizes its ORF3 (VP2) genetically. The status of BNoV should be continuously evaluated in Egypt for effective prevention and control.
1 Introduction
Calf diarrhea is considered a significant contributor to economic losses worldwide. Bovine norovirus (BNoV) is an important cause of diarrhea in cattle [Citation1] either alone or with other viral entero-pathogens. BNoV was found in the feces of both diarrheic and healthy animals [Citation2–Citation4]. On experimental infection, gnotobiotic calves infected with BNoV showed various degrees of diarrhea and anorexia as a result of gastroenteritis [Citation5,Citation6].
The BNoV is a small, non-enveloped, and positive sense single-stranded RNA virus, which belongs to the family Caliciviridae. The viral genome is 7.3 to 7.5 kb in size and consists of three open reading frames (ORF1, ORF2, and ORF3), which encode for large polyprotein, major capsid protein (VP1) and small basic protein (VP2), respectively [Citation7]. Based on the phylogeny of deduced amino acids (aa) of VP1 protein, noroviruses are classified into seven genogroups (GI–GVII); GI, GII, and GIV infect humans while GIII infects bovine and sheep [Citation8]. Each genogroup is further divided into several genotypes. For example, GIII is divided into GIII.1 (Bo/Jena/1980/DE) and GIII.2 (Bo/Newbury-2/1976/UK) originally discovered in Germany [Citation9] and England [Citation10], respectively. Ovine norovirus and recombinant BNoV strains represent newer genotypes in GIII [Citation11,Citation12].
The reverse transcription-polymerase chain reaction (RT-PCR) is commonly used for norovirus detection. The RT-PCR usually targets the RNA dependent RNA polymerase (BNoV-RdRp) gene because it contains highly conserved regions. For phylogenetic classification, RT-PCR targets the BNoV-VP1 gene. Recently, several recombination events have been detected at the ORF1/ORF2 junction, thus imposing a new genotyping strategy within the Norovirus genus. It was reported that GIII.P1/GIII.2 had a GIII.1-related RdRp and a GIII.2-related VP1 while GIII.P2/GIII.1 had a GIII.2-related RdRp and a GIII.1-related VP1. This indicates that both BNoV-RdRp and BNoV-VP1 should be determined for phylogenetic purposes [Citation13].
In noroviruses, VP2 is not essential for virion production but it is mainly responsible for virion stability and genome encapsidation. Besides, VP2 interacts with VP1 and, in turn, enhances the expression of capsid proteins [Citation14]. At the molecular level, VP2 is highly variable and can express different types of mutations [Citation15,Citation16]. Little is known about BNoV-VP2 due to the limited number of available sequences.
In Egypt, few data are available on the newly emerging enteric viruses of cattle, including BNoV. Only one study described BNoV infection in two cattle farms in Egypt [Citation17]. The present study was conducted to (i) report the existence of BNoV among diarrheic calves in Sharkia province of Egypt and (ii) to study the phylogenesis of these BNoV strains on the basis of VP2 gene.
2 Materials and methods
2.1 Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
2.2 Samples and RNA extraction
Twenty-nine fecal specimens were collected aseptically from sporadic cases of diarrheic calves in Sharkia province of Egypt between late 2016 and early 2017. Calves had diarrhea, dehydration, and weakness with different degrees. The age of calves ranged from 3 weeks to 3 months. No deaths were observed at the time of sample collection. According to the case history, animals were not vaccinated against locally-identified enteric viruses, including bovine rotavirus (BRV) and bovine viral diarrhea virus (BVDV). All samples were diluted in phosphate buffer saline (10×) and centrifuged at 2500×g for 10 min. The viral RNA was extracted from the supernatant fluid of each sample (140 µL) by following the manufacturer's instructions of the QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA, USA).
2.3 Reverse transcription-polymerase chain reaction (RT-PCR)
Initially, the one-step RT-PCR (QIAGEN, Valencia, CA, USA) was carried out to detect the BNoV RNA in tested fecal samples by targeting a conserved area in BNoV-RdRp gene (4543–5074) using the primer pair CBECu-F/R [Citation18]. The PCR cyclic conditions were 50 °C/30 min for reverse transcription, 95 °C/15 min for PCR activation, and 35 cycles of 94 °C/1 min for denaturation, 51 °C/45 s for annealing, and 72 °C/1 min for elongation followed by a final cycle of extension at 72 °C/10 min. Then, another primer pair (BNoV-VP2-F/R) was designed to flank the BNoV-VP2 gene from base 6432 to base 7257 (826 bp) in the positive samples. The design of these primers was made based on the VP2 region of the GenBank reference sequence (Newbury2/UK/AF097917). The previous PCR cycle was used except for annealing that was changed to 55 °C for 1 min. The PCR reactions were 25 μL in volume (1 μL enzyme mix, 12.5 μL buffer, 1 μL forward primer, 1 μL reverse primer, 2.5 μL of extracted RNA, and 7 μL RNase-free water). In negative controls, a 9.5 μL of RNase-free water was added with no RNA. The PCR amplicons were allowed to run through agarose gel by electrophoresis and then were visualized on UV transilluminator to visualize the band size. The oligonucleotide primers used in this study are listed in .
Table 1 The oligonucleotide primers used in this study.
2.4 Sanger sequencing
The PCR products of BNoV-VP2 were purified using a QIAquick PCR purification kit (QIAGEN, Valencia, CA, USA) and sequenced, directly in both directions using the same primers previously used in RT-PCR, at the University of Minnesota Genomics Center (UMGC). Using Sequencher 5.1 software (http://genecodes.com/), contigs were generated using forward and reverse sequences. The obtained sequences were further identified by online nucleotide BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST).
2.5 Sequence and phylogenetic analysis
The study sequences had the accession numbers of MF784575 and MF784576 in GenBank. Using the MEGA 6 software [Citation19], nucleotide (nt) sequences of BNoV-VP2 were comparatively aligned to reference strains using the Clustal W option. The phylogenetic tree was constructed using the neighbor-joining statistical method (1.000 bootstrap replicates). The identities of nt and aa were calculated using the p-distance method. The aa were deduced from BNoV-VP2 nt sequences and analyzed using the DNASTAR Lasergene 7.2 software [Citation20].
3 Results
3.1 Molecular detection of BNoV in diarrheic fecal samples
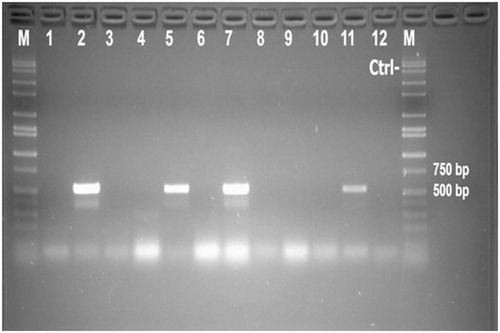
Using the RT-PCR with BNoV-RdRp specific primers, 8 of 29 fecal samples (27.6%) were confirmed positive for BNoV-RNA. Positive samples showed specific bands of 532 bp on 1.5% agarose gel under UV rays (). The previous step aimed to rule out the negative samples for BNoV as the RdRp is highly conserved and commonly used for BNoV detection. Furthermore, positive RNA samples were retested by RT-PCR but with BNoV-VP2 targeting primers in order to characterize the VP2 gene molecularly. The designed primers annealed successively to their sites; nt 6432–6449 on the plus strand for the forward primer and nt 7257–7237 on the minus strand for the reverse primer and the expected bands (826 bp) were demonstrated on the gel. Samples tested negative for BRV, BVDV, and bovine coronavirus (data not shown).
3.2 Sequence and phylogenetic analysis of study BNoV-VP2 strains
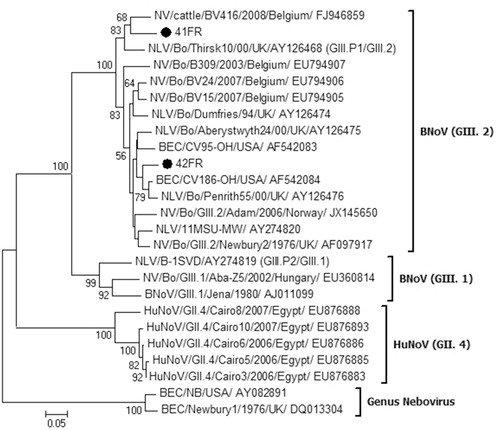
The molecular characters of BNoV-VP2 strains were investigated by sequencing an 826 bp region. Homogenous hits were obtained from GenBank upon blasting and used in the phylogenetic tree, which revealed that study strains (41FR and 42FR) clustered within BNoV-GIII.2 genotype (Newbury2-like). However, they were separated from each other within the same major cluster. The 41FR strain grouped with BV416/Belgium and Thirsk10/UK (recombinant GIII.P1/GIII.2 strains) but the 42FR strain grouped with CV186-OH/USA and Penrith55/UK ().
At nt and aa levels, 41FR and 42FR had identities of 81.8% and 88.5%, respectively. The 41FR was closely related to BV416/Belgium (84.3% nt identity and 89.7% aa identity) while 42FR was more related to CV186-OH/USA (identities of 89.5% and 89.9% at nt and aa levels, respectively). In relation to Newbury2/UK/AF097917, 41FR and 42FR shared nt identities of 79.1–86.5% and aa identities of 86.7–88.9% ().
Table 2 The nucleotide (nt) and amino acid (aa) identities between study strains (41FR and 42FR) and reference BNoV-GIII.2 strains.
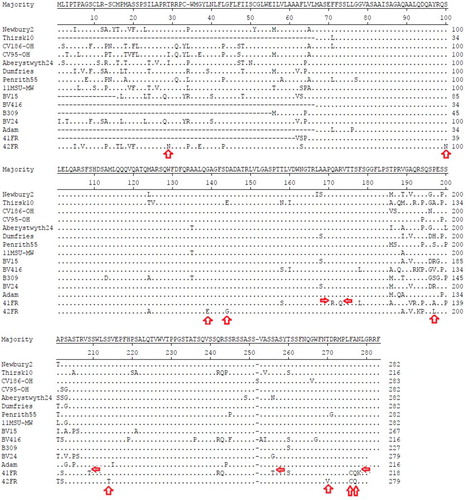
The VP2 regions from BNoV-GIII.2 reference strains (n = 13) were obtained from blast hits. Their deduced aa sequences were aligned with those of 41FR and 42FR to detect substitution mutations in our strains. Newbury2/UK/AF097917 was selected as a reference base for building the alignment (282 aa in length). The consensus sequence for VP2 proteins was determined based on the major aa present at each position in the 15 (13 reference and 2 study) strains. Overall, study strains (41FR and 42FR) had 41 substitutions compared to the consensus sequence. Among them, 14 substitutions were found to be unique to our strains. Substitutions at positions 171 (Q to R), 173 (R to Q), 209 (S to T), 255 (S to T) and 277 (N to K) were found only in 41FR. Meanwhile, mutations at positions 29 (T to N), 100 (S to N), 139 (G to E), 144 (D to G), 197 (P to L), 214 (S to T) and 269 (T to V) were found only in 42FR. Both 41FR and 42FR shared two unique substitutions at positions 275 (F to C) and 276 (A to Q) as shown in .
4 Discussion
Noroviruses are one of the leading causes of human and animal gastroenteritis. The circulation of BNoV has been reported in Europe, America, Asia, and Africa [Citation21]. However, BNoV is usually not included in diagnostic algorithms of calf diarrhea. Accordingly, its impact on livestock industries is unknown. In Africa, the reports of BNoV-infections came only from Egypt and Tunisia [Citation17,Citation22].
The results of this study confirm those of Mohamed et al. [Citation17], who found the detection rates of BNoV at 24% in Egyptian cattle as compared to 27.6% in this study. The results of both studies collectively indicate the endemic nature of BNoV infection in Egyptian cattle. The detected BNoV strains were genetically-related to BNoV-GIII.2 genotype (Newbury2-like), either based on BNoV-RdRp [Citation17] or based on BNoV-VP2 (this study). BNoV-GIII.2 has a worldwide distribution, unlike BNoV-GIII.1 [Citation23]. Calves under the age of one month were the most affected with BNoV (data not shown).
To address BNoV-VP2 molecularly, representative strains from GenBank (n = 23) were used to construct a neighbor-joining phylogenetic tree (). BNoV strains were phylogenetically-distinct from human norovirus (HuNoV) strains, excluding the possibility of interspecies transmission. Although being classified as BNoV-GIII.2, study strains (41FR and 42FR) were separated from each other. Studies on HuNoVs in Egypt suggested that ORF3 (VP2) followed ORF2 (VP1) in its genetic and phylogenetic characters. However, due to the slightly different topologies between ORF2 and ORF3, BNoV-VP2 may have distinct evolutionary strategies [Citation24]. Interestingly, the 41FR was related to recombinant GIII.P1/GIII.2 strains (BV416/Belgium and Thirsk10/UK), raising questions about the presence of intra-genotypic recombinations. We are planning for complete genome sequencing of 41FR strain to detect possible recombination breakpoints.
Fourteen unique aa substitution mutations were detected in both strains (). Neither insertions nor deletions were observed. Such variations may play a role in classifying closely-related BNoV strains. Study strains shared identities of 81.8% and 88.5% at nt and aa, respectively. The nt and aa identities () further confirm the clustering made by the phylogenetic tree. It is expected for norovirus capsid proteins, particularly the VP2, to exhibit broad sequence variability [Citation15]. The hypervariability of VP2 sequences was assumed to be functionally-related to those occurring in the P2 domain of VP1 protein [Citation25] and this may affect the viral pathogenesis.
5 Conclusions
The study reported the presence of BNoV-GIII.2 for the second time in Egypt as well as the phylogenetic properties of BNoV-VP2. It is concluded that BNoV is circulating among diarrheic cattle of Egypt. The genetic diversity of BNoV strains should be monitored to prevent future economic losses related to the virus.
Competing interests
The content of this paper is not related personally or financially to other people or organizations.
Author contributions
FM provided the study specimens (sample source) and initiated their preparation. SG supervised the project. FM, SG conceived and planned the experiments, contributed to reagents/materials/analysis tools. FM, GK, SG contributed to the interpretation of the results. FM carried out the experiments and took the lead in writing the manuscript in consultation with others.
Acknowledgments
The authors thank the Ministry of Higher Education and Scientific Research (MHESR) in Egypt for providing a visiting grant to FFM.
Notes
Peer review under responsibility of Faculty of Veterinary Medicine, Cairo University.
References
- A.ScipioniA.MauroyJ.VinjeE.ThiryAnimal norovirusesVet J17820083245
- W.H.Van der PoelR.van der HeideF.VerschoorH.GelderblomJ.VinjéM.P.KoopmansEpidemiology of Norwalk-like virus infections in cattle in The NetherlandsVet Microbiol922003297309
- E.JorM.MyrmelC.M.JonassenSYBR Green based real-time RT-PCR assay for detection and genotype prediction of bovine noroviruses and assessment of clinical significance in NorwayJ Virol Methods169201017
- B.Di MartinoF.Di ProfioE.Di FeliceI.MelegariC.CeciA.MauroyGenetic heterogeneity of bovine noroviruses in ItalyArch Virol159201427172722
- P.H.OttoI.N.ClarkeP.R.LambdenO.SalimJ.ReetzE.M.Liebler-TenorioInfection of calves with bovine norovirus GIII. 1 strain Jena virus: an experimental model to study the pathogenesis of norovirus infectionJ Virol8520111201312021
- K.JungK.A.ScheuerZ.ZhangQ.WangL.J.SaifPathogenesis of GIII. 2 bovine norovirus, CV186-OH/00/US strain in gnotobiotic calvesVet Microbiol1682014202207
- S.L.OliverE.AsobayireA.CharpilienneJ.CohenJ.C.BridgerComplete genomic characterization and antigenic relatedness of genogroup III, genotype 2 bovine norovirusesArch Virol1522007257272
- K.Y.GreenCaliciviridae: the norovirusesD.M.KnipeP.M.HowleyFields virologySixth Edvol. 22013Lippincott Williams & WilkinsPhiladelphia, USA582608
- H.GüntherP.OttoDiarrhea in young calves. 7.“ Zackenvirus” (Jena agent 117/80)–a new diarrhea pathogen in calvesArch Exp Veterinarmed411987934938
- G.N.WoodeJ.C.BridgerIsolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calvesJ Med Microbiol111978441452
- S.WolfW.WilliamsonJ.HewittS.LinM.Rivera-AbanA.BallMolecular detection of norovirus in sheep and pigs in New Zealand farmsVet Microbiol1332009184189
- R.A.BullM.M.TanakaP.A.WhiteNorovirus recombinationJ Gen Virol88200733473359
- F.FerragutC.G.VegaA.MauroyN.Conceição-NetoM.ZellerE.HeylenMolecular detection of bovine Noroviruses in Argentinean dairy calves: Circulation of a tentative new genotypeInfect Genet Evol402016144150
- S.VongpunsawadB.V.PrasadM.K.EstesNorwalk virus minor capsid protein VP2 associates within the VP1 shell domainJ Virol87201348184825
- E.L.SeahI.C.GunesekereJ.A.MarshallP.J.WrightVariation in ORF3 of genogroup 2 Norwalk-like virusesArch Virol144199910071014
- K.BokE.J.AbenteM.Realpe-QuinteroT.MitraS.V.SosnovtsevA.Z.KapikianEvolutionary dynamics of GII. 4 noroviruses over a 34-year periodJ Virol8320091189011901
- F.F.MohamedS.M.MansourI.E.El-ArabyS.K.MorS.M.GoyalMolecular detection of enteric viruses from diarrheic calves in EgyptArch Virol1622017129137
- J.R.SmileyA.E.HoetM.TråvénH.TsunemitsuL.J.SaifReverse transcription-PCR assays for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human calicivirusesJ Clin Microbiol41200330893099
- K.TamuraG.StecherD.PetersonA.FilipskiS.KumarMEGA6: molecular evolutionary genetics analysis version 6.0Mol Biol Evol30201327252729
- T.G.BurlandDNASTAR’s Lasergene sequence analysis softwareMethods Mol Biol13220007191
- E.Di FeliceA.MauroyF.Dal PozzoD.ThiryC.CeciB.Di MartinoBovine noroviruses: a missing component of calf diarrhoea diagnosisVet J20720165362
- M.Hassine-ZaafraneJ.KaplonK.Sdiri-LouliziZ.AouniP.PothierM.AouniMolecular prevalence of bovine noroviruses and neboviruses detected in central-eastern TunisiaArch Virol157201215991604
- A.MauroyA.ScipioniE.MathijsD.ZiantG.DaubeE.ThiryGenetic and evolutionary perspectives on genogroup III, genotype 2 bovine norovirusesArch Virol15920143949
- A.H.KamelM.A.AliH.G.El-NadyA.De RougemontP.PothierG.BelliotPredominance and circulation of enteric viruses in the region of Greater Cairo, EgyptJ Clin Microbiol47200910371045
- M.C.W.ChanN.LeeW.S.HoC.O.K.LawT.C.K.LauS.K.W.TsuiCovariation of major and minor viral capsid proteins in norovirus genogroup II genotype 4 strainsJ Virol86201212271232