Abstract
Melioidosis, caused by Burkholderia pseudomallei, is an emerging infectious disease with an expanding geographical distribution. Although assessment of the environmental load of B. pseudomallei is important for risk assessment in humans or animals in endemic areas, traditional methods of bacterial culture for isolation have low sensitivities and are labor-intensive. Using a specific polymerase chain reaction (PCR) assay targeting a Tat domain protein in comparison with a bacterial culture method, we examined the prevalence of B. pseudomallei in soil samples from an oceanarium in Hong Kong where captive marine mammals and birds have contracted melioidosis. Among 1420 soil samples collected from various sites in the oceanarium over a 15-month period, B. pseudomallei was detected in nine (0.6%) soil samples using bacterial culture, whereas it was detected in 96 (6.8%) soil samples using the specific PCR assay confirmed by sequencing. The PCR-positive samples were detected during various months, with higher detection rates observed during summer months. Positive PCR detection was significantly correlated with ambient temperature (P<0.0001) and relative humidity (P=0.011) but not with daily rainfall (P=0.241) or a recent typhoon (P=0.787). PCR-positive samples were obtained from all sampling locations, with the highest detection rate in the valley. Our results suggest that B. pseudomallei is prevalent and endemic in the oceanarium. The present PCR assay is more sensitive than the bacterial culture method, and it may be used to help better assess the transmission of melioidosis and to design infection control measures for captive animals in this unique and understudied environment.
Introduction
Burkholderia pseudomallei is an emerging, highly pathogenic, gram-negative beta-proteobacterium responsible for melioidosis, a potentially serious and fatal disease often manifesting as community-acquired pneumonia and sepsis. Although melioidosis is mainly endemic in Southeast Asia and northern Australia, the disease has been increasingly reported in countries outside the Asia-Pacific region, including India,Citation1,Citation2 Mauritius,Citation3 South, Central and North America,Citation4,Citation5,Citation6 and West and East Africa,Citation7,Citation8 suggesting an expanding geographical distribution and/or awareness. The illness can present as an acute, subacute or chronic process. Disease manifestations range from subclinical infection localized abscesses to severe pneumonia and fulminant sepsis, with case fatality rates of up to 19% in endemic areas.Citation9 The incubation period of melioidosis also varies widely from 2 days to 26 years.Citation10 Diagnosis of melioidosis can be difficult, as the bacterium may not be readily isolated from clinical specimens. Moreover, even with positive cultures, commercial bacterial identification kits often fail to distinguish between B. pseudomallei and closely related species such as B. thailandensis and B. cepacia complex.Citation11 Nevertheless, the advent of new molecular techniques has enabled the development of improved methods for more accurate species identification.Citation12,Citation13,Citation14,Citation15,Citation16,Citation17,Citation18 Treatment of melioidosis may be difficult, as B. pseudomallei is often resistant to multiple antibiotics, and a prolonged course of antibiotics is required to prevent disease relapse.Citation13,Citation19 Unfortunately, in many of the endemic areas and countries, diagnostic and therapeutic resources are limited, hindering efforts to better assess the disease burden and improve treatment outcomes.
B. pseudomallei is a natural saprophyte that can be isolated from soil, groundwater, stagnant streams, rice paddies and ponds, which, together, are the major natural reservoirs of the bacteria.Citation20,Citation21 Although its epidemiology and route of transmission are not fully understood, melioidosis is believed to be acquired through environmental contact with contaminated soil and contaminated water by percutaneous inoculation, inhalation of aerosols or ingestion.Citation22 Owing to its high mortality rates, antibiotic resistance and possible transmission by aerosols, B. pseudomallei is considered a potential agent of biological warfare and has been classified as a category B bioterrorism agent by the Center for Disease Control (Atlanta, GA, USA; http://www.bt.cdc.gov/agent/agentlist-category.asp). Human cases are often spatially and temporally clustered and may follow heavy rains and winds with exposure to soil and water.Citation23,Citation24 B. pseudomallei also causes melioidosis in a wide range of animals in endemic areas.Citation25 In Hong Kong, melioidosis is an endemic disease not only in humans but also in captive marine mammals and birds, including bottlenose dolphins, California sea lions, pilot whales and zebra doves.Citation12 Strains of B. pseudomallei with closely related genotypes have been isolated from soil and water collected in the neighborhood of infected animals.Citation12 However, the environmental distribution of B. pseudomallei in Hong Kong is poorly understood.
Assessment of the environmental load of B. pseudomallei may help in estimating the disease risk and deciding possible preventive measures in endemic areas. Moreover, knowledge of its environmental distribution, in relation to specific habitats and factors such as climate change, is important for understanding the epidemiology of melioidosis. However, the gold standard for B. pseudomallei detection in environmental samples is culture, which lacks sensitivity and is time-consuming. Molecular methods based on detection of bacterial nucleic acids have the potential to overcome the problems of culture-based methods. Therefore, different polymerase chain reaction (PCR) assays have been reported to detect B. pseudomallei.Citation26,Citation27,Citation28,Citation29 To detect B. pseudomallei DNA from environmental samples, a highly specific gene target is essential, as B. pseudomallei is phylogenetically closely related to B. thailandensis and other Burkolderia species that may be found in the same environment. Using a pan-genomic analysis approach in gene target selection, we previously developed a novel and specific PCR assay targeting a Tat domain protein for the identification and detection of B. pseudomallei from soil and simulated sputum samples.Citation30 In this study, we examined the prevalence of B. pseudomallei in soil samples from an oceanarium in Hong Kong where captive animals have been infected with melioidosis,Citation12 and we evaluated the sensitivity of the PCR assay compared to culture-based detection methods.
MATERIALS AND METHODS
Soil samples
Soil samples were prospectively collected each month from various sites in the oceanarium from June 2010 to August 2011, a period encompassing two wet seasons and one dry season. Briefly, a standard soil sampling technique was used,Citation31 with approximately 200 g of soil collected from a depth of 20–30 cm. Soil samples were sealed in plastic containers at the ambient temperature and immediately transported to the laboratory for enrichment and bacterial culture.
Culture, isolation and identification of B. pseudomallei
Bacterial culture and isolation of B. pseudomallei were performed according to previously published protocols with modifications.Citation32 Briefly, 100 g of each soil sample was mechanically homogenized with 100 mL of purified water. The mixture was left to settle at 25 °C overnight, and 1 mL of the resulting soil supernatant was collected for enrichment in 9 mL of modified Ashdown’s broth containing 10 g/L tryptic soy broth (Oxoid, Basingstoke, Hampshire, UK), 40 mL/L glycerol (UltraPure, Waltham, Massachusetts, USA), 5 mg/L crystal violet (Sigma-Aldrich, St. Louis, MO, USA) and 1 million international units (MIU)/L colomycin (Forest Laboratories UK Ltd., Dartford, Kent, UK) and in 9 mL Galimand’s broth supplemented with 1 MIU/L colomycin (Forest Laboratories UK). The cultures were incubated aerobically at 42 °C for 10 days. Ten microliters of each enriched culture supernatant was plated on Ashdown's agar, containing 10 g/L trypticase soy broth (Oxoid), 40 mL/L glycerol (Ultrapure), 5 mg/L 0.1% crystal violet (Sigma-Aldrich), 50 mg/L neutral red, 5 mg/L gentamicin (Gibco, Waltham, Massachusetts, USA) and 15 g/L agar, and incubated aerobically at 42 °C for 48 h. The colonies gown on Ashdown’s agar plates were screened for B. pseudomallei morphotypes. Suspected B. pseudomallei isolates were phenotypically identified by the API 20NE system (bioMérieux Vitek, Hazelwood, MO, USA) and Vitek 2 system (bioMérieux Vitek) supplemented by conventional biochemical methods.
PCR detection of B. pseudomallei
One milliliter of enriched soil culture supernatant from Ashdown’s broth was harvested for bacterial DNA extraction using the QIAamp DNA mini kit (QIAgen, Hilden, Germany) according to the manufacturer’s instructions. A single-target PCR assay for B. pseudomallei was performed using B. pseudomallei-specific primers targeting a 189-bp fragment of a specific gene that encodes a Tat domain protein (locus BPSS0658 in the B. pseudomallei K96243 reference genome); the protocol was modified from our previously described multiplex PCR assay.Citation30 The PCR mixture (20 µL) contained purified DNA extract (1.0 µL) as template, 1.0 M betaine monohydrate (Fluka BioChemika, Steinheim, Germany), 0.5 µM primers (LPW13372: 5′-CAA GAA CGG TTT ATG CG-3′ and LPW13373: 5′-GAA GTG ATC CAT CAA ATG TC-3′), 2.0 µL 10× PCR buffer II, 2.5 mM MgCl2, 200 µM of each dNTPs (GeneAmp, Applied Biosystems, Waltham, Massachusetts, USA) and 1.0 U Taq polymerase (AmpliTaq Gold; Applied Biosystems, Waltham, Massachusetts, USA). Thermal cycling was performed in an automated thermocycler (Veriti 96-well fast thermal cycler; Applied Biosystems, Waltham, Massachusetts, USA) with a hot-start at 95 °C for 10 min; 10 touch-down cycles of 95 °C for 30 s, annealing for 1.5 min at temperatures decreasing from 60 °C to 51 °C (with 1.0 °C decremental steps) and 72 °C for 1 min; 30 cycles of 95 °C for 30 s, 50 °C for 1.5 min and 72 °C for 1 min; and a final extension at 72 °C for 10 min. Five microliters of each amplified product was electrophoresed in 2.5% (w/v) agarose gel with a molecular size marker (GeneRuler 50 bp DNA Ladder; Fermentas, Pittsburgh, PA, USA) in parallel. Electrophoresis in Tris-borate-EDTA buffer was performed at 100 V for 45 min. The gel was stained with ethidium bromide (0.5 µg/mL) for 25 min, rinsed and photographed under ultraviolet light illumination. Standard precautions were taken to avoid PCR contamination, and no false-positive was observed in negative controls.
The PCR products were gel-purified using the QIAquick PCR gel extraction kit (QIAgen). Both strands of the PCR products were sequenced with an ABI 3130xl Genetic Analyzer according to the manufacturer’s instructions (Applied Biosystems) using primers specific to each PCR product. The obtained DNA sequences were analyzed using a BLASTx search of the in-house Burkholderia pan-genome databases and by a BLASTn search against the NCBI online nucleotide collection (nr/nt) database to confirm their identities. The specificity of the PCR assay has been previously confirmed using pure isolates of closely related bacterial species, including B. pseudomallei, B. thailandensis and B. cepacia.Citation30
Statistical analyses
Correlation of PCR detection with ambient temperature, relative humidity, rainfall and recent typhoon was performed using logistic regression. P<0.05 was regarded as statistically significant (IBM SPSS Statistics 19, Armonk, New York, USA).
RESULTS
Culture and isolation of B. pseudomallei from soil samples
A total of 1420 soil samples were collected from the oceanarium, comprising 90–120 samples per month during the 15-month study period. The samples were collected from various sites at three different locations, including lowland, headland and valley areas situated at different altitudes ( and Supplementary Figure S1). Soil samples from the different locations all consisted of decomposed granite. Among the 1420 samples, nine (0.6%) samples were positive for B. pseudomallei by bacterial culture, ranging from 0% to 5.6% of samples taken in a given month (Figure ). The positive isolates were detected in August 2010 (four isolates) in a valley area and in November 2010 (five isolates) in both lowland and headland areas facing the sea (Figure and ). No positive cultures could be recovered during the other months.
Figure 1 (A) Detection of B. pseudomallei from soil samples by PCR and culture during the study period and in relation to ambient temperature and rainfall. (B) Monthly number of melioidosis cases in captive animals in the oceanarium between 2002 and 2011. Data showed the cumulative cases in the respective months over a 10-year period (2002-2011).
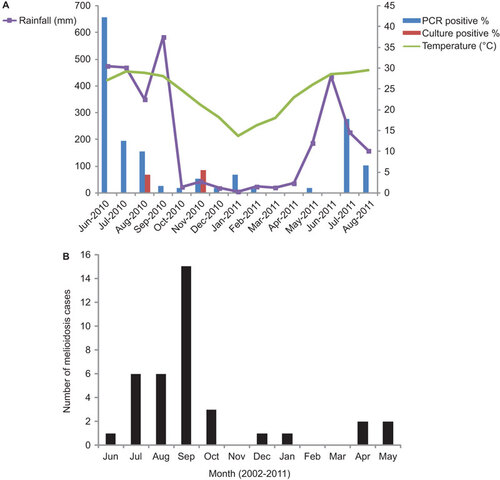
Table 1 Distribution of B. pseudomallei in different locations of the oceanarium
PCR for detection of B. pseudomallei from soil samples
Using a specific PCR assay targeting a B. pseudomallei-specific Tat domain protein-encoding gene, 96 (6.8%) of the 1420 samples showed positive bands of approximately 189 bp, corresponding to the expected PCR product size, by gel electrophoresis (Supplementary Figure S2). The positive detection rates ranged from 0% to 42.2% of samples taken in a given month (Figure ). DNA sequencing of the positive PCR products confirmed that they originated from the target locus, with 100% nucleotide identities to the corresponding gene fragment of B. pseudomallei strain K96243 (GenBank accession NO BX571966). No positive reactions were found for any negative controls during the same PCR runs, thus excluding PCR contamination. The PCR-positive samples were detected during various months throughout the study period, with the exceptions of March, April and June 2011. Higher detection rates were observed during the summer months, when both ambient temperature and relative humidity were high (such as June–August 2010 and July–August 2011); the highest detection rate was recorded in June 2010 (42.2%) (Figure and ). By logistic regression, significant correlations were demonstrated between positive detection and ambient temperature (P<0.0001) or relative humidity (P=0.011) on the day of sampling, but not between positive detection and daily rainfall (P=0.241) or a typhoon within the 7 days prior to sampling (P=0.787). PCR-positive samples were derived from all the three sampling locations of the oceanarium, including the lowland, headland and valley areas, with the highest detection rate in the valley ( and Supplementary Figure S1).
Table 2 Seasonal distribution of B. pseudomallei in the oceanarium
DISCUSSION
The present study confirms that B. pseudomallei is endemic in the soil environment of the present oceanarium, where captive animals have been infected by the bacterium. Moreover, the present PCR assay is more sensitive than the culture method for detection of B. pseudomallei in soil samples. B. pseudomallei has been found in soil samples from endemic areas, including Thailand, southern China, Taiwan and northern Australia.Citation21,Citation26,Citation27,Citation33,Citation34 Although most previous studies have relied on the culture of viable bacteria from soil samples, there has been increasing interest in developing molecular detection methods. Although multiple PCR assays have been developed for such purposes, some assays still lack sensitivity or specificity.Citation28,Citation29 Moreover, few studies have directly compared the sensitivities of PCR and culture methods. In one study, a quantitative PCR (qPCR) detection assay was developed and validated using 40 soil samples from northeast Thailand.Citation26 Among 26 of 40 soil samples that tested positive for B. pseudomallei by culture, all were also positive by qPCR.Citation26 Moreover, qPCR detected the bacterium in seven additional samples that were negative by culture. In another study from northern Australia, a real-time PCR assay was evaluated using enriched soil samples.Citation27 In addition to the 13 of 104 soil samples testing positive by both culture and qPCR, seven further samples were positive by qPCR but not by culture.Citation27 The present PCR assay also achieves higher sensitivity, with a detection rate >10-fold higher than culture methods for detection of B. pseudomallei from soil samples. However, it is difficult to compare the sensitivities of the different PCR assays used in different studies because different methods and gene targets were employed. Instead of the direct/enriched soil samples used in the two previous studies, the PCR assay in this study was performed using the enriched culture supernatant as the template to avoid the problem of PCR inhibitors that are often encountered in soils.Citation26,Citation27 Similar methods using enriched culture supernatant for PCR have also been reported for groundwater samples.Citation21 This assay may offer a cheaper alternative to real-time PCR methods, which may not be available in some endemic areas or countries. The superiority of PCR-based assays over culture-based detection can be explained by the problems associated with the culture and isolation of B. pseudomallei. Cultivation depends on efficient selection of B. pseudomallei over other, often more rapidly growing environmental bacteria on the chosen selective media. Moreover, culture can only detect cultivable bacterial cells; it will not detect potentially viable but non-culturable cells, which may underestimate the B. pseudomallei bacterial load in environmental habitats. Although a positive PCR result does not imply the presence of viable bacteria, it represents a sensitive surrogate marker for the presence of B. pseudomallei in the environment. Nevertheless, as enriched culture supernatant was used in this study, a PCR-positive result in our soil samples may imply the presence of viable bacteria. Further studies on the application of the present and other molecular method-based assays are required to assess their usefulness for detecting B. pseudomallei in different environmental samples.
The Tat domain protein represents a sensitive and specific alternative target for PCR detection of B. pseudomallei. In previous studies using molecular detection of B. pseudomallei from environmental samples, the type III secretion system (TTSS) and, less commonly, the flagellin and BPSS1187 genes have been used as specific gene targets.Citation21,Citation26,Citation27,Citation33,Citation34 In studies from both Thailand and Australia, a 115-bp fragment of the single-copy TTSS1 gene was used as the gene target for amplification.Citation26,Citation27 TTSS1 has been found to be ubiquitously present in B. pseudomallei but not in close relatives such as B. thailandensis or B. mallei.Citation35 In our previous study, different gene targets specific to B. pseudomallei, B. thailandensis and B. cepacia complex (the Tat-domain protein, a 70-kDa protein and a 12-kDa protein, respectively) were selected using a pan-genomic analysis approach.Citation30 Based on the three gene targets, a multiplex PCR assay was developed and found to be sensitive and specific for detection of the respective bacteria in simulated sputum samples.Citation30 A pilot study using 60 soil samples allowed the detection of B. pseudomallei in 19 (31.6%) samples and B. cepacia complex in 29 (48.3%) samples, with codetection of both bacteria in four (6.7%) samples. The apparently higher detection rate of B. pseudomallei in the pilot study than in the present study is likely due to the use of soil samples collected during the peak season, as opposed to the samples in the present study collected across different seasons. A single-target PCR assay based on the Tat-domain protein, found only in B. pseudomallei and not in B. thailandensis or B. cepacia complex, was chosen in place of the multiplex PCR assay for detection of B. pseudomallei in the present study. This strategy was designed to minimize the chance of false-negatives, which can occur in the multiplex assay as a result of interactions from the presence of B. cepacia DNA in the same soil sample. The results confirmed that the single PCR assay targeting the Tat-domain protein-encoding gene is specific for detecting B. pseudomallei and is more sensitive than culture methods.
Environmental detection of B. pseudomallei is important for disease anticipation and infection control measures to combat melioidosis in endemic areas, such as in the captive animals of the present oceanarium. The detection of B. pseudomallei in soil is believed to be related to the risk of developing melioidosis. For example, higher bacterial counts of B. pseudomallei from soil in the northeastern region of Thailand than in the central region may contribute to the higher incidence of reported melioidosis cases in the former region.Citation20 In a recent report from northern Australia, a B. pseudomallei strain cultured from an athlete with cutaneous melioidosis was identical by multilocus sequence typing and multilocus variable-number tandem repeat analysis to an isolate recovered from the soil at the location on the sports field where he was injured.Citation24 Such findings may alert clinicians to consider the possibility of melioidosis in persons from endemic areas with abrasion injuries that involve contact with soil.Citation24 However, as culture methods are more labor intensive and less sensitive, molecular detection should be the first-line method for environmental detection of B. pseudomallei; it can be supplemented by culture-based methods for verification of positive results. Despite B. pseudomallei having been discovered nearly a century ago,Citation36 its geographical distribution and ecology in its natural habitat remains poorly understood. In China, a few reports have revealed the presence of B. pseudomallei in soil or water from southern coastal provinces, including Hainan, Guangdong and Guangxi.Citation34,Citation37 The present assay serves as an alternative, sensitive molecular detection method to explore the soil distribution of B. pseudomallei in other potential endemic areas.
The higher PCR detection rate of B. pseudomallei in soil samples during summertime and the positive correlation of detection with ambient temperature and relative humidity may explain the seasonality of melioidosis in both humans and animals in Hong Kong, where sporadic cases or small outbreaks are mainly observed during summer. In our oceanarium, the seasonality of melioidosis cases among the captive animals from 2002 to 2011 also correlated with the monthly trend of PCR-positive soil samples in this study, with higher incidence during summer than winter months (Figure ). Although correlation with humidity has not been reported previously, studies have associated human melioidosis with rainfall, suggesting that the infection may result from acute exposure to the organism in the soil and water.Citation9,Citation23,Citation28,Citation32,Citation38,Citation39 A case of fulminant infection was reported following exposure to stagnant floodwater in India.Citation1 Two fatal cases of human melioidosis have also been reported in Queensland, Australia, with disease onset preceded by unseasonal heavy rainfall.Citation39 A subsequent study in Queensland demonstrated that the timing and location of 47 melioidosis cases was generally correlated with rainfall across northern Australia, with a case cluster associated with post-cyclonic flooding.Citation40 In another study from northern Australia involving 318 cases, rainfall in the 14 days before hospital admission was found to be an independent risk factor for pneumonia, septic shock and death, suggesting that heavy monsoonal rains and winds may cause a shift toward inhalation of B. pseudomallei.Citation23 In northeast Thailand, the disease also showed a strong correlation with rainfall, and adults exposed to soil and water at work, such as rice farmers, had an increased risk of melioidosis.Citation38 Similar positive linear associations have also been demonstrated between monthly rainfall and melioidosis cases and/or deaths in Malaysia and India.Citation41,Citation42 Although we did not find significant correlation between positive PCR detection and daily rainfall or recent typhoons, these factors may have caused delayed effects on bacterial load, or other factors may have had a greater impact on the ecology of B. pseudomallei in the unique environment of our oceanarium. Further studies are warranted to understand the role of climate changes, such as global warming, in the emergence of melioidosis in different endemic and non-endemic areas.
Supplementary Figure s1
Download PDF (144.1 KB)Supplementary Figure s2
Download PDF (171.4 KB)PCR and sequencing work is partly supported by the HKSAR Research Fund for the Control of Infectious Diseases (Commissioned Study HK-09-01-11) of the Health, Welfare and Food Bureau. This work was also supported by the Strategic Research Theme Fund and University Development Fund, The University of Hong Kong; the Shaw Foundation; and by a donation from Ms Eunice Lam. Work on bacterial cultures is funded by the oceanarium.
Note: Supplementary Information for this article can be found on Emerging Microbes and Infections' website (http://www.nature.com/emi/).
- Anuradha K, Meena AK, Lakshmi V.Isolation of Burkholderia pseudomallei from a case of septicaemia—a case report. Indian J Med Microbiol2003;21: 129–132.
- Saravu K, Mukhopadhyay C, Vishwanath S et al.Melioidosis in southern India: epidemiological and clinical profile. Southeast Asian J Trop Med Public Health2010;41: 401–409.
- Issack MI, Bundhun CD, Gokhool H.Melioidosis in mauritius. Emerg Infect Dis2005;11: 139–140.
- Inglis TJ, Rolim DB, Sousa AD.Melioidosis in the Americas. Am J Trop Med Hyg2006;75: 947–954.
- Rolim DB, Vilar DC, Sousa AQ et al.Melioidosis, northeastern Brazil. Emerg Infect Dis2005;11: 1458–1460.
- Stewart T, Engelthaler DM, Blaney DD et al.Epidemiology and investigation of melioidosis, Southern Arizona. Emerg Infect Dis2011;17: 1286–1288.
- Salam AP, Khan N, Malnick H, Kenna DT, Dance DA, Klein JL.Melioidosis acquired by traveler to Nigeria. Emerg Infect Dis2011;17: 1296–1298.
- Cuadros J, Gil H, Miguel JD et al.Case report: melioidosis imported from West Africa to Europe. Am J Trop Med Hyg2011;85: 282–284.
- Currie BJ, Fisher DA, Howard DM et al.Endemic melioidosis in tropical northern Australia: a 10-year prospective study and review of the literature. Clin Infect Dis2000;31: 981–986.
- Mays EE, Rickets EA.Melioidosis: recrudescence associated with bronchogenic carcinoma twenty-six years following initial geographic exposure. Chest1975;68: 261.
- Weissert C, Dollenmaier G, Rafeiner P, Riehm J, Schultze D.Burkholderia pseudomallei misidentified by automated system. Emerg Infect Dis2009;15: 1799–1801.
- Godoy D, Randle G, Simpson AJ et al.Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol2003;41: 2068–2079.
- Woo PC, Lau SK, Woo GK et al.Seronegative bacteremic melioidosis caused by Burkholderia pseudomallei with ambiguous biochemical profile: clinical importance of accurate identification by 16S rRNA gene and groEL gene sequencing. J Clin Microbiol2003;41: 3973–2977.
- Lau SK, Tang BS, Curreem SO et al.Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of Burkholderia pseudomallei: importance of expanding databases with pathogens endemic to different localities. J Clin Microbiol2012;50: 3142–3143.
- Inglis TJ, Healy PE, Fremlin LJ, Golledge CL.Use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis for rapid confirmation of Burkholderia pseudomallei in septicemic melioidosis. Am J Trop Med Hyg2012;86: 1039–1042.
- Payne GW, Vandamme P, Morgan SH et al.Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl Environ Microbiol2005;71: 3917–3927.
- Schmoock G, Ehricht R, Melzer F et al.DNA microarray-based detection and identification of Burkholderia mallei, Burkholderia pseudomallei and Burkholderia spp. Mol Cell Probes2009;23: 178–187.
- Woo PC, Woo GK, Lau SK, Wong SS, Yuen K.Single gene target bacterial identification. groEL gene sequencing for discriminating clinical isolates of Burkholderia pseudomallei and Burkholderia thailandensis. Diagn Microbiol Infect Dis2002;44: 143–149.
- Cheng AC, Currie BJ.Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev2005;18: 383–416.
- Smith MD, Wuthiekanun V, Walsh AL, White NJ.Quantitative recovery of Burkholderia pseudomallei from soil in Thailand. Trans R Soc Trop Med Hyg1995;89: 488–490.
- Baker A, Tahani D, Gardiner C, Bristow KL, Greenhill AR, Warner J.Groundwater seeps facilitate exposure to Burkholderia pseudomallei. Appl Environ Microbiol2011;77: 7243–7246.
- Currie BJ, Mayo M, Anstey NM, Donohoe P, Haase A, Kemp DJ.A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am J Trop Med Hyg2001;65: 177–179.
- Currie BJ, Jacups SP.Intensity of rainfall and severity of melioidosis, Australia. Emerg Infect Dis2003;9: 1538–1542.
- Hill AA, Mayo M, Kaestli M et al.Melioidosis as a consequence of sporting activity. Am J Trop Med Hyg2013;89: 365–366.
- Dance DA.Melioidosis. Curr Opin Infect Dis2002;15: 127–132.
- Trung TT, Hetzer A, Göhler A et al.Highly sensitive direct detection and quantification of Burkholderia pseudomallei bacteria in environmental soil samples by using real-time PCR. Appl Environ Microbiol2011;77: 6486–6494.
- Kaestli M, Mayo M, Harrington G et al.Sensitive and specific molecular detection of Burkholderia pseudomallei, the causative agent of melioidosis, in the soil of tropical northern Australia. Appl Environ Microbiol2007;73: 6891–6897.
- Brook MD, Currie B, Desmarchelier PM.Isolation and identification of Burkholderia pseudomallei from soil using selective culture techniques and the polymerase chain reaction. J Appl Microbiol1997;82: 589–596.
- Sonthayanon P, Krasao P, Wuthiekanun V, Panyim S, Tungpradabkul S.A simple method to detect and differentiate Burkholderia pseudomallei and Burkholderia thailandensis using specific flagellin gene primers. Mol Cell Probes2002;16: 217–222.
- Ho CC, Lau CC, Martelli P et al.Novel pan-genomic analysis approach in target selection for multiplex PCR identification and detection of Burkholderia pseudomallei, Burkholderia thailandensis, and Burkholderia cepacia complex species: a proof-of-concept study. J Clin Microbiol2011;49: 814–821.
- Limmathurotsakul D, Wuthiekanun V, Chantratita N et al.Burkholderia pseudomallei is spatially distributed in soil in Northeast Thailand. PLoS Negl Trop Dis2010;4: e694.
- Wuthiekanun V, Smith MD, Dance DA, White NJ.Isolation of Pseudomonas pseudomallei from soil in north-eastern Thailand. Trans R Soc Trop Med Hyg1995;89: 41–43.
- Su HP, Yang HW, Chen YL et al.Prevalence of melioidosis in the Er-Ren River Basin, Taiwan: implications for transmission. J Clin Microbiol2007;45: 2599–2603.
- Ma G, Zheng D, Cai Q, Yuan Z.Prevalence of Burkholderia pseudomallei in Guangxi, China. Epidemiol Infect2010;138: 37–39.
- Rainbow L, Hart CA, Winstanley C.Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis and B. mallei. J Med Microbiol2002;51: 374–384.
- Woods D, Sokol P.Proteobacteria: alpha and beta subclasses.In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed.) The prokaryotes.New York: Springer Press, 2006: 848–860.
- Yang S, Tong S, Lu Z.Geographical distribution of Pseudomonas pseudomallei in China. Southeast Asian J Trop Med Public Health1995;26: 636–638.
- Suputtamongkol Y, Hall AJ, Dance DA et al.The epidemiology of melioidosis in Ubon Ratchatani, northeast Thailand. Int J Epidemiol1994;23: 1082–1090.
- Scott IA, Bell AM, Staines DR.Fatal human melioidosis in south-eastern Queensland. Med J Aust1997;166: 197–199.
- Cheng AC, Hanna JN, Norton R et al.Melioidosis in northern Australia, 2001–02. Commun Dis Intell Q Rep2003;27: 272–277.
- Hassan MR, Pani SP, Peng NP et al.Incidence, risk factors and clinical epidemiology of melioidosis: a complex socio-ecological emerging infectious disease in the Alor Setar region of Kedah, Malaysia. BMC Infect Dis2010;10: 302.
- Vidyalakshmi K, Lipika S, Vishal S, Damodar S, Chakrapani M.Emerging clinico-epidemiological trends in melioidosis: analysis of 95 cases from western coastal India. Int J Infect Dis2012;16: e491–e497.
