Abstract
Recent years have seen a global and rapid resurgence of fungal diseases with direct impact on biodiversity and local extinctions of amphibian, coral, or bat populations. Despite similar evidence of population extinction in European fish populations and the associated risk of food aquaculture due to the emerging rosette agent Sphaerothecum destruens, an emerging infectious eukaryotic intracellular pathogen on the fungal–animal boundary, our understanding of current threats remained limited. Long-term monitoring of population decline for the 8-year post-introduction of the fungal pathogen was coupled with seasonal molecular analyses of the 18S rDNA and histological work of native fish species organs. A phylogenetic relationship between the existing EU and US strains using the ribosomal internal transcribed spacer sequences was also carried out. Here, we provide evidence that this emerging parasite has now been introduced via Pseudorasbora parva to sea bass farms, an industry that represents over 400 M annually in a Mediterranean region that is already economically vulnerable. We also provide for the first time evidence linking S. destruens to disease and severe declines in International Union for Conservation of Nature threatened European endemic freshwater fishes (i.e. 80% to 90 % mortalities). Our findings are thus of major economic and conservation importance.
Introduction
Recent years have seen a global and rapid resurgence of fungal diseases with direct impact on biodiversity with local extinctions of amphibian, coral, and bat populationsCitation1,Citation2,Citation3 as well as declines in crop production leading to a loss of food for 600–4200 million mouths per annum.Citation4 Despite similar evidence of population extinction in European fish populations due to an emerging fungal diseaseCitation5 and the associated risk of food aquaculture,Citation6,Citation7,Citation8 our understanding of global distribution of fish fungal pathogens today remains limited.Citation9,Citation10,Citation11 This limitation which is characterized by a severe under reporting from the scientific community, stems from a lack of diagnostic power in aquatic systems, the chronic nature of these diseases, which spread slowly through populations and their lack of obvious external pathological signs in the hosts during the initial phase of infection.Citation12,Citation13
In the early sixties, building on privileged commercial partnerships between China and a number of former countries from the Union of Soviet Socialist Republics, a small cyprinid freshwater fish, the topmouth gudgeon Pseudorasbora parva, was accidentally introduced to a range of river catchments surrounding the Black Sea.Citation14 These initial introductionsCitation15 have since fuelled one of the fastest fish invasions in the world at a rate of about five new countries invaded each decade.Citation14 Recently, this invasive fish species has been identified as a healthy carrier of the rosette agent Sphareothecum destruens, a generalist pathogen on the animal–fungal boundary.Citation16 This novel pathogen caused mass mortality in a range of fish species including wild and farmed Chinook salmon in California where it caused mass mortality of smolt (>90%).Citation7,Citation8 Although there is evidence of isolation between S. destruens’ strain found in the United States of America (USA) and the one found in Europe and that the detected hosts are from different families (e.g. salmonids, cyprinids), further analyses have shown that it was in fact the same fungal pathogen species responsible for severe mortalities on both continents.Citation17
Since these initial discoveriesCitation5 and in spite of evidence of P. parva status as a healthy carrierCitation18,Citation19 and further experimental evidence of the severe impact of this fungal pathogen on a range of European fish species,Citation20 no monitoring of this disease distribution within P. parva populations has yet been put in place with the exception of the Netherlands.Citation19 This painfully highlights the key limitations as often seen with other pandemicsCitation21,Citation22 for experimental evidence on emerging pathogens to influence environmental agencies and policy makers responsible for the setting up of monitoring programs and disease prevention in aquaculture.Citation11,Citation23 In particular in the Mediterranean region, where there is a high level of fish endemicity for which 60% of these endemic species are classified as vulnerable, endangered, or critically endangered by the International Union for Conservation of Nature (IUCN).Citation24 In Europe, two Chinese lineages of the healthy carrier P. parva have been introduced. One originating north of the Yangtze river and which colonized the whole western part of Europe including Southern England where S. destruens was initially discovered and one originating south of the Yangtze river which colonized Bulgaria, Turkey, and Armenia.Citation15
Here, for the first time, we want to show that the emergence of S. destruens is associated with severe declines in endemic fish species in the wild and that this pathogen has now escaped the freshwater ecosystem to include marine species of high commercial value such as European sea bass Dicentrarchus labrax. With around 120 000 tonnes of D. labrax produced annually across the Mediterranean region and for a value of about €700 M, the presence of S. destruens would represent a major risk to European aquaculture production. Finally, we also looked for potential levels of geographical isolation across the different S. destruens isolates in support of the role of ancestral association between P. parva and S. destruens during the rapid European invasion.
Materials and methods
Sampling
A set of endemic freshwater fish species (Oxynoemacheilus sp. not yet described, Petroleuciscus smyrnaeus, Squalius fellowesii), a marine native species (D. labrax) and two nonnative species (P. parva, Lepomis gibbosus) have been sampled monthly by backpack electrofishing (model SAMUS 725 MP) in the Sarıçay stream, Muğla, Turkey at three locations in the main river channel (site 1 37°20′37.15″N, 27°43′43.11″E; site 2 37°19′43.10″N, 27°42′45.73″E; site 3 37°18′8.87″N, 27°42′44.87″E). The sampling was started in July 2009 and ended in April 2013. All electrofishing sampling consisted of a team of three people, one operator (the same each time) with the anode and two others with hand nets walking through the river from downstream to upstream for a set time (circa 30 min). The catch per unit was then calculated as species abundance per unit of time (min). This catchment corresponds to the edge of P. parva invasion in southeastern Europe, which was first detected in 2006 from a founder population with a genetic lineage originating south of the Yangtze river, China. Some specimens were brought back to the laboratory alive, where they were euthanized with an overdose of 2-phenoxyethanol and severance of the spine.
Diagnostic work-up
A total of 112 fish were euthanased in the laboratory with an overdose of methane tricaine sulfonate (MS-222, Sigma-Aldrich, Saint Louis Missouri, USA) and processed. Postmortem examination included a note on general external appearance and the presence of any obvious lesions or abnormalities as well as a record of species, size, location, and date. Then, the operculum was removed to observe the gills and a ventrolateral opening was made in the body of the fish along the flank below the lateral line, curving the cut down to the anus. Kidney, gonad, spleen, and liver tissues (n = 448) were then collected and preserved half in 10% formalin and the other half directly extracted for molecular analysis. Using standard histological procedures and sections stained with haematoxylin and eosin (H&E), slides were examined under a microscope (40–100× magnification).
DNA extraction and polymerase chain reaction (PCR) amplification
The collected tissues were homogenized by pestle in lysis buffer and incubated at 56 °C overnight and DNA extracted using commercial kits (Thermo Scientific GeneJET Genomic DNA purification kit and Qiagen Dneasy Blood & Ttissue kit). All samples were subjected to nested PCR assay using the S. destruens 18S rDNA primers.Citation20 Cycling conditions consisted of an initial denaturation cycle at 95 °C for 5 min followed by 35 cycles of 45 s at 95 °C, 45 s at 60 °C, and 45 s at 72 °C. A final elongation step at 72 °C was performed for 7 min. For first amplification forward (Sd-1F) and reverse (Sd-1R) primer was 5′-CGA CTT TTC GGA AGG GAT GTA TT 3′ and 5′-AGT CCC AAA CTC GAC GCA CAC T-3′. For second amplification forward (Sd-2F) and reverse (Sd-2R) primer was 5′-CCC TCG GTT TCT TGG TGA TTC ATA ATA ACT-3′ and 5′-CTC GTC GGG GCA AAC ACC TC-3′. PCR products were electrophoresed with 1.5% agarose gel including 1 µL EtBr in TAE buffer and visualized under ultraviolet light analyzer. PCR products were sequenced by REFGEN laboratories. DNA sequences were compared to sequences of BLAST database. An S. destruens DNA sample from the United Kingdom (UK) strain was used as a positive control and amplified by PCR in the same conditions.
As PCR tests are so sensitive, all PCR work was done with a negative control to address the measure of potential contamination. Some samples were also blind checked at Bournemouth University laboratory, which has ten years of expertise in S. destruens detection using molecular analyses.
Internal transcribed spacer (ITS) sequence analysis
The ITS sequence used for comparisons originated from isolates obtained from three North American outbreaks: (i) Chinook salmon in Washington (accession number: FJ440706); (ii) Atlantic salmon in California (accession number: FJ440708); (iii) winter-run Chinook salmon in California (accession number: FJ440702). The sources of these three American cultures are deposited in the American Type Culture Collection (ATCC) under accession numbers 50643, ATCC 50644, and ATCC 50615, respectively. In addition, the ITS sequences for the UK isolates are deposited in GenBankTM under the accession numbers FJ440702–FJ440710.Citation17
A nested PCR was performed to amplify the ITS1 region of the S. destruens isolate from Turkey. PCR reactions were prepared in 50 μL vols. with Go-Taq Flexi DNA polymerase 5× Master Mix (Promega, Madison, WI, USA), to a final concentration of 1×, containing 1.5 mM MgCl2, 0.2 mM dNTP, and 1.25 units Taq polymerase. PCR primers were used at a concentration of 0.25 μM each. The first reaction utilized the forward primer Sdes2F (5′-CTT CGG ATT GGC CCT GTA C-3′) and the NC13R (5′-GCT GCG TTC TTC ATC GAT-3′).Citation25 Five microliters of template DNA were added to the first PCR reaction and a PCR was run on the C1000 thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA), as follows: 95 °C for 3 min, 35 × [95 °C for 30 s, 52 °C for 45 s, 62 °C for 90 s], with a final extension at 62 °C for 7 min.
Two microliters of the first round PCR were then used to perform the second round PCR. The second round PCR used the primers Sdes2F and SD-ITSR1 (5′-CGA TGC ACG AGC CAA GAG-3′) yielding a PCR product of approximately 765 bp.
Products were visualized on a 1.0% agarose gel containing 0.1 lg mL−1 ethidium bromide run at 125 V for 30 min. Products were excised from the gel and purified using the Wizard SV Gel and PCR clean-up system (Promega). Purified fragments were cloned using the Topo TA cloning kit (Invitrogen, Life Technologies, Paisley, United Kingdom) and then sequenced with M13 forward and reverse. At least three clones were sequenced for each sample. Sequences were assembled manually using BioEditCitation26 and identity verified by alignment to existing S. destruens ITS1 sequences and GenBank by performing a BLAST search.Citation27 The amplified ITS1 sequence from the Turkish isolate was deposited in GenBank, accession numbers KT361608. For phylogenetic analysis, ITS1 sequences were aligned with Clustal X version 1 and examined by eye in BioEdit. We tested for the best substitution model using MEGA 5Citation28 and performed the phylogenetic analysis using maximum likelihood and the Tamura 3-parameter modelCitation28 in MEGA 5 with bootstrap support calculated using 1000 replicates. The phylogenetic tree is unrooted as no appropriate outgroup is available due to the ITS sequence from other mesomycetozoean species being so different that no confidence can be placed on the homology of most nucleotide positions. Genetic distances between isolates were calculated using DNADIST in BioEdit.Citation26
Results
Prevalence of S. destruens in fish population
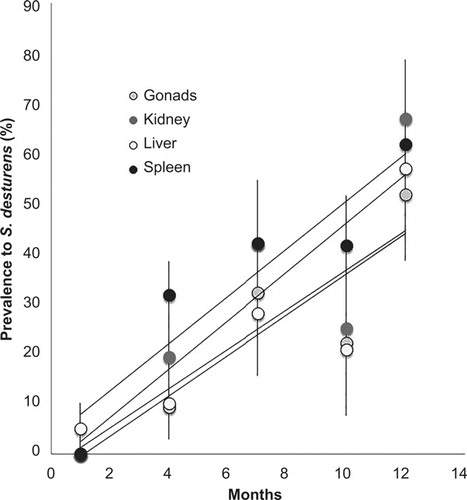
All fish species tested were infected by S. destruens with prevalences ranging from 25% to 100% (). Prevalence is high despite the relatively low number of individuals randomly subsampled, although these samples sizes are comparable to other epidemiological studies on wild populations (). Vulnerable endemic species S. fellowesii, Oxynoemacheilus sp. (in the process of being described) and P. smyrnaeus show high prevalence of the pathogen throughout most seasons. Even the nonnative L. gibbosus, a centrarchid species showed prevalence as high as 86%. This is the first time that a centrarchid species has been found to be sensitive to S. destruens. Due to the commercial value of sea bass D. labrax only three specimens were sampled and showed a prevalence of 67%, which correspond to the values found in highly sensitive hosts.
Table 1 Seasonal prevalence of S. destruens per organ for wild caught fish in 2012 and 2013 in Sarıçay stream detected by PCR
All organs tested kidney, liver, spleen, and ovary consistently tested positive to S. destruens, which confirmed the non-organ specificity of the pathogen. The organ the most affected across all freshwater species and seasons was consistently the kidney (66, 8%) followed by the spleen (61, 6%) and the liver (56, 8%); the spores can infect further tissues or be excreted through the bile, urine, gut epithelium, and seminal and ovarian fluids. Similar prevalence is observed in the marine L. labrax with the highest prevalence in kidney and spleen.
A clear temporal increase in the level of infections with the freshwater fish community is observed in all four organs tested ().
Mortalities
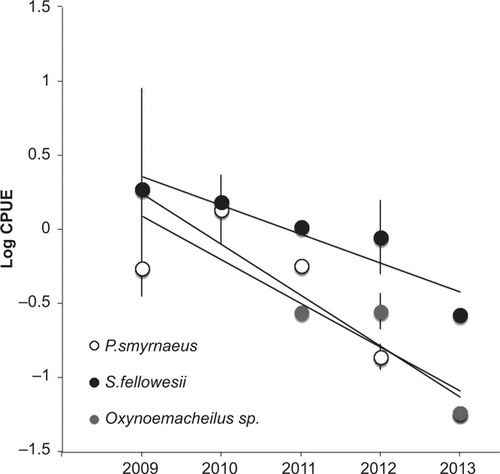
All three endemic species of high conservation status that have cohabited in the wild with P. parva have severely declined since 2009 with rates comprised of 80% to 90% (). In light of increased temporal prevalence of S. destruens in the community during this period of time, it is likely that this vortex of extinction characterizes the emergence of this infectious disease in the wild. Although all fish species were severely affected, Oxynoemacheilus sp., which has not yet been described as a species, showed the lowest abundance, which puts this species in critical danger of extinction.
Figure 2 Decline of three endemic species cohabiting in the wild since P. parva introduction in that catchment. Catch per unit efforts (CPUE) is calculated from species abundance per unit of time (min) and averaged over three locations on the main river channel. Standard errors are also included. Regression lines were fitted (R2 = 0.85, 0.74, 0.72 for S. fellowesii, Oxynoemacheilus sp., and P. smyrnaeus respectively).
Pathology
The histological work-up did not present specifically novel findings and associated pathology in the various tissues of the different species was similar to that published previously (Supplementary Figure S1).Citation5,Citation6,Citation8,Citation12,Citation13,Citation20 Parasitized fish did not show any internal gross signs of disease and did not appear emaciated. However in D. labrax, hemorrhages on the liver and white nodules on the kidneys were observed similarly to what has previously been found on Atlantic salmon Salmo salar, another example of a marine host.Citation6 Sphaerothecum destruens infection was systemic in all species and was observed in all vital organs, including the gonads, kidney, liver, and spleen (). S. destruens is deeply eosinophilic with H&E. Here we observed the disseminated form of the disease where the agent is widely dispersed throughout the fish (Supplementary Figure S1a) with little host cell immune response. A nodular form of the disease also exists and is characterized by host cell immune response and the formation of distinct granulomas in visceral organs, which has not been observed here. The disseminated form is expected to indicate a host more susceptible to S. destruens while granuloma formations are considered more able to contain the infection. Various isolated stages of spore development were observed, with sizes ranging between 4 μm and 7 μm in diameter. Most stages appeared to be intracellular but were also occasionally observed extracellularly (Supplementary Figure S1). In D. labrax’s liver, some inflammatory responses were present with phagocytic cells containing the spores and some lymphocytic infiltration in the parenchyma. However, in most tissues across all species, the parasites were present with minimal host cell response, most likely due to subclinical fish being collected.Citation11 S. destruens’ life stages include spherical intra-cytoplasmic spore stages (Supplementary Figures S1a and S1b). Spores replicate asexually through fission and can infect epithelial, mesenchymal, and hematopoietic cells, eventually causing cell death.
Phylogeny
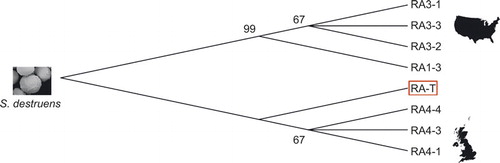
PCR amplification of 620 bp of the ribosomal ITS1 identified the Turkish isolate to be closely related to the UK isolate, but the two are not identical. Analysis of the ITS1 of the European isolates, when compared with the variability of the first reported isolates in the USA indicates genetic differences observed between geographically distinct isolates () highlighting a lack of intermixing between European and North American S. destruens populations. The genetic distance between the Turkish S. destruens isolate differs and the UK isolate was 1.7% compared to 2.5% with the US isolate. In the aligned sequences there are seven polynucleotide indels, which distinguish the North American and European (UK Turkey) isolates whereas one polynucleotide indel occurs between the European isolates. A further eight single and dinucleotide indels and seven-point mutations separate the two European isolates suggesting that there is no intermixing between these two isolates.
Figure 3 Phylogenetic tree resulting from maximum likelihood method based on the Tamura 3-parameter modelCitation29 analysis of the ribosomal ITS1 sequence from S. destruens. The bootstrap consensus tree inferred from 1000 replicatesCitation30 and bootstrap values exceeding 50% are shown at nodes. The phylogenetic tree was produced using MEGA 5.Citation28 Isolate (and clone) origin, designations and GenBank accession numbers are: RA1-3 (FJ440707.1); RA3-1 (FJ440708.1); RA3-2 (FJ440709.1); RA3-3 (FJ440710.1); RA4-1 (FJ440702.1); RA4-3 (FJ440703.1); RA4-4 (FJ440704.1); and RA-T (to be deposited).
Discussion
Our results show how the introduction of a freshwater fish in Europe 50 years ago could rapidly lead to a pan-continental disease risk to European endemic species in a region where 56% of endemic fish species are threatened and 18% critically endangeredCitation24 and spill over to marine species. Here we found that in the wild all fish species that cohabited with P. parva were highly infected with the pathogen S. destruens. We also show a severe decline of endemic fish species 3-year post P. parva introduction where catch per unit effort dropped by 80% to 90%. These levels are comparable to the declines due to S. destruens observed in the USA and the UK in salmonids and cyprinids respectively. Of course, correlation is not causation and there are indeed very few direct measures of animal disease outbreaks in the wild following pathogen introductions and the few iconic ones have always tried to link population decline and infectious pathogen introduction.Citation1,Citation2,Citation3,Citation4,Citation21 Here, in addition there have been several experimental trials that have demonstrated the direct link between S. destruens introduction and mortalities (i.e. causation) as well as large-scale mesocosm ones. Thus, the conclusions do not rely on population decline alone but (i) on pathogen emergence in the populations since carrier introduction in this catchment; (ii) on quantitative measures of disease prevalence using specific PCR pre- and post-start of decline; (iii) on histological work-up showing disease in the declining species (Supplementary Figure S1). As everything else in the catchment remained constant during this long time series, there are limited additional ways to show historical wild emergence of disease in aquatic speciesCitation11 and as such these results are stronger than the initial reports on chytrid emergence in amphibians or Aphanomyces astaci (crayfish plague) in native crayfish populations for example.
The results highlight that the farming of marine fish in the brackish part of estuaries represents a risk of contamination to pathogens initially carried by freshwater hosts, a risk that has so far been overlooked.Citation31 Despite previous cases of severe S. destruens emergence in Californian Chinook and Atlantic salmon farms (c. >90% mortalities),Citation32 it was believed due to the anadromous nature of these species that the contact with the pathogen occurred in freshwater.Citation8,Citation32 This understanding was further reinforced as zoosporulation of S. destruens only occurred in fresh water.Citation8,Citation33
It is now apparent that the risk of spill over from freshwater hosts to marine ones exists with levels of prevalence considerably higher than previously found in wild Chinook salmon Oncorhynchus tshawytscha (32%)Citation12 and Leucaspius delineatus (2%). The location of D. labrax farming in the brackish part of estuaries represents a front of contact with invasive infectious P. parva populations, a species that has been previously shown to be tolerant to brackish conditions.Citation34 It also shows that contaminations are likely to have occurred via direct predation of infected P. parva or infected native species, as free-living zoospores are solely produced in freshwater.Citation8,Citation33 Currently, the extent of the risk posed by S. destruens to D. labrax farming remains unquantified but severe mortalities as observed previously in smolt cages in US farms is not to be ruled out.
In addition, the phylogenetic analysis of S. destruens has revealed a higher level of relatedness between the UK and the Turkey strains when compared to the North American ones (). The reason may be found in the invasion history of the carrier that displayed two distinct routes of invasion in Europe. The first route started from an initial introduction into Romania, which colonized the Danube river and via fish movements from Germany ended up in the southern part of the UK where S. destruens was first discovered. The second route corresponds to an initial introduction that took place in Bulgaria and which spread southward to colonize Turkish rivers.Citation15 The original P. parva source populations of S. destruens in the UK, belongs to a native lineage that is distributed in China north of the Yangtze river, whilst those found in Turkey correspond to a lineage found south of this river. The relatedness of the UK and Turkish S. destruens strains with different host lineages is a strong indication, as initially hypothesized,Citation5,Citation14,Citation17 that S. destruens and P. parva association in Europe dates back to the original introductions. However, the fact that UK and Turkish ITS sequences are not identical may result from an ancient host–pathogen association that goes back prior to the segregation of the Chinese northern and southern P. parva lineages.Citation15
The results have to be further confirmed but an ancient coevolution between P. parva and S. destruens is likely to show direct implications on the transmission and virulence of the pathogen and may have facilitated the rapid establishment of P. parva during the invasion process, particularly due to the non-specificity of the pathogen (e.g. cyprinids, centrarchids, salmonids). A rapid screening of P. parva populations in Europe and across the native range for the presence of S. destruens and the phylogeny of the various strains should help clarify the origin of S. destruens. It will also help us to check if the contamination of salmonid populations on the west coast of the USA by S. destruensCitation7,Citation8 finds its origin in the North Pacific or across the bearing sea where Chinese, Russian, Japanese, and North American salmonids cohabitCitation35,Citation36 and where Asian salmonids have been cohabiting with P. parva populations since the seventies due to fish translocations in China.Citation15
Supplementary Figure S1
Download PDF (1.2 MB)Note: Supplementary Information for this article can be found on Emerging Microbes and Infections' website (http://www.nature.com/emi/)
- Cheng TL, Rovito SM, Wake DB, Vredenburg VT.Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proc Natl Acad Sci USA 2011; 108: 9502–9507.
- Kim K, Harvell CD.The rise and fall of a six-year coral-fungal epizootic. Am Nat 2004; 164: S52–S63.
- Frick WF, Pollock JF, Hicks AC et al.An emerging disease causes regional population collapse of a common North American bat species. Science 2010; 329: 679–682.
- Fisher MC, Henk DA, Briggs CJ et al.Emerging fungal threats to animal, plant and ecosystem health. Nature 2012; 484: 186–194.
- Gozlan RE, St-Hilaire S, Feist SW et al.Disease threats on European fish. Nature 2005; 435: 1045–1046.
- Paley RK, Andreou D, Bateman KS, Feist SW.Isolation and culture of Sphaerothecum destruens from Sunbleak (Leucaspius delineatus) in the UK and pathogenicity experiments in Atlantic salmon (Salmo salar). Parasitology 2012; 139: 904–914.
- Harrell LW, Elston RA, Scott TM, Wilkinson MT.A significant new systemic disease of net-pen reared Chinook salmon (Oncorhynchus tshawytscha) brood stock. Aquaculture 1986; 55: 249–262.
- Arkush KD, Mendoza L, Adkison MA, Hedrick RP.Observations on the life stages of Sphaerothecum destruens n.g., n. sp., a Mesomycetozoean fish pathogen formally referred to as the Rosette Agent. J Eukaryot Microbiol 2003; 50: 430–438.
- Rowley JJL, Gleason FH, Andreou D et al.Impact of mesomycetozoean parasites in amphibian and freshwater fish populations. Fungal Ecol Rev 2013; 27: 100–111.
- Gozlan RE, Marshall WL, Lilje O et al.Current ecological understanding of fungal-like pathogens of fish: what lies beneath? Front Microbiol 2014; 5: 62.
- Gozlan RE.Monitoring fungal infections in fish. Nature 2012; 285: 446.
- Arkush KD, Frasca SJr, Hedrick RP.Pathology Associated with the Rosette Agent, A Systemic Protist Infecting Salmonid Fishes. J Aquat Anim Health 1998; 10: 1–11.
- Andreou D, Feist SW, Stone D et al.First occurrence and associated pathology of Sphaerothecum destruens in cyprinids. J Aquat Dis Organ 2011; 95: 145–151.
- Gozlan RE, Andreou D, Asaeda T et al.Pan-continental invasion of Pseudorasbora parva: towards a better understanding of freshwater fish invasions. Fish & Fisheries 2010; 11: 315–340.
- Simon A, Britton JR, Gozlan RE et al.Invasive cyprinid fish in Europe originate from the single introduction of an admixed source population followed by long-distance dispersal. PLoS One 2011; 6: e18560.
- Mendoza L, Taylor JW, Ajello L.The class Mesomycetozoea: A group of microorganisms at the animal-fungal boundary. Annu Rev Microbiol 2002; 56: 315–344.
- Gozlan RE, Whipps C, Andreou D, Arkush K.Characterisation and geographical isolation of Sphaerothecum destruens in Europe. Int J Parasitol 2009; 39: 1055–1058.
- Andreou D, Hussey M, Griffiths SW, Gozlan RE.Influence of host reproductive state on Sphaerothecum destruens prevalence and infection level. Parasitology 2011; 138: 26–34.
- Spikmans F, van Tongeren T, van Alen TA et al.High prevalence of the parasite Sphaerothecum destruens in the invasive topmouth gudgeon Pseudorasbora parva in the Netherlands, a potential threat to native freshwater fish. Aquat Inv 2013; 8: 355–360.
- Andreou D, Arkush KD, Guégan J-F, Gozlan RE.Introduced pathogens and native freshwater biodiversity: A case study of Sphaerothecum destruens. PLoS One 2012; 7: e36998
- Vincent ER.Whirling disease and wild trout. Fisheries 1996; 21: 1–88.
- Clifton-Hadley RS, Bucke D, Richards RH.Proliferative kidney disease of salmonid fish: a review J Fish Dis 1984; 7: 363–377.
- Gozlan RE, Burnard D, Andreou D, Britton JR.Understanding the threats posed by non-native species: Public vs. conservation managers. Plos One.2013; 8: e53200.
- Smith KG, Darwall WRT. The status and distribution of freshwater fish endemic to the Mediterranean Basin.Cambridge: IUCN, 2006.
- Chilton NB, Gasser RB.Sequence differences in the internal transcribed spacers of rDNA among four species of hookworm (Ancylostomatoidea: Ancylostoma). Int J Para 1999; 29: 1971–1977.
- Hall TA.BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41: 95–98.
- Thompson JD, Gibson TJ, Plewniak F et al.The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997; 25: 4876–4882.
- Tamura K, Peterson D, Peterson N et al.MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28: 2731–2739.
- Tamura K.Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol 1992; 9: 678–687.
- Felsenstein J.Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39: 783–791.
- Peeler EJ, Oidtmann BC, Midtlyng PJ et al.Non-native aquatic animals introductions have driven disease emergence in Europe. Biol Inv 2010; 13: 1291–1303.
- Kent ML.Disease of seawater netpen-reared salmonid fishes in the Pacific Northwest. Nanaimo. Can Spec Publ Fish Aquat Sci 1992; 116: 1–76 .
- Andreou D, Paley R, Gozlan RE.Temperature influence on production and longevity of Sphaerothecum destruens’ zoospores. Parasitology 2009; 95: 1539–1541.
- Scott DM, Wilson RW, Brown JA.Can sunbleak Leucaspius delineatus or topmouth gudgeon Pseudorasbora parva disperse through saline waters? J Fish Biol 2007; 71: 70–86.
- Shigehiko U, Sato S, Crane PA et al.Stock-specific ocean distribution and migration of Chum Salmon in the Bering Sea and North Pacific Ocean. N Pac An Fish Com Bul 2009; 5: 131–146.
- Sato S, Kato M, Morita K, Urawa S.Stock-specific summertime distribution of immature Chum Salmon in the Bering Sea as inferred from SNP markers. N Pac An Fish Com Tech Rep 2012; 8: 50–51.