Abstract
Fusarium is a rapidly emerging, multidrug-resistant genus of fungal opportunists that was first identified in 1958 and is presently recognized in numerous cases of fusariosis each year. The authors examined trends in global Fusarium distribution, clinical presentation and prevalence since 1958 with the assumption that their distributions in each region had remained unaltered. The phylogeny and epidemiology of 127 geographically diverse isolates, representing 26 Fusarium species, were evaluated using partial sequences of the RPB2 and TEF1 genes, and compared with AFLP fingerprinting data. The molecular data of the Fusarium species were compared with archived data, which enabled the interpretation of hundreds of cases published in the literature. Our findings indicate that fusariosis is globally distributed with a focus in (sub)tropical areas. Considerable species diversity has been observed; genotypic features did not reveal any clustering with either the clinical data or environmental origins. This study suggests that infections with Fusarium species might be truly opportunistic. The three most common species are F. falciforme and F. keratoplasticum (members of F. solani species complex), followed by F. oxysporum (F. oxysporum species complex).
INTRODUCTION
Fusarium infections are a major challenge with respect to the diagnosis and treatment, especially in neutropenic patients. Disseminated infections may be fatal and are a considerable source of increased healthcare costs. A major area of concern is the intrinsic resistance to a broad range of antifungals,Citation1 which is a characteristic of Fusarium. During the past decade, the F. solani complex has received special interest because of the increasing numbers of infections worldwide.Citation2 More than 300 cases of Fusarium keratitis were associated with contaminated contact lens cleaning solution, causing outbreaks between 2005 and 2007, where members of the F. solani species complex played a major role.Citation3 Furthermore, reservoirs of infectious Fusarium species in hospital environments, especially plumbing and water systems, have been reported.Citation4
Although human fusariosis was only recognized since the late 1950s and endemic areas are mostly located in tropical and subtropical countries,Citation5 their global significance has only recently come into focus within the past three decades. Etiological agents differ in antifungal susceptibility,Citation6 virulence profiles, geographic distribution, ecological niches, life cycle, host and mycotoxin production.Citation7 Although agents of fusariosis are mostly environmental,Citation8 the potential of nosocomial transmission has recently been raised,Citation9 especially with reference to the high mortality rate of ~90% in patients with prolonged and severe neutropenia.Citation10
The burden of disease has not been established, but numerous clinical case series and case reports provide an estimate of the magnitude of the problem. Most published studies have focused on prevalence in single healthcare centers.Citation10, Citation11, Citation12, Citation13, Citation14, Citation15, Citation16 Nucci et al.Citation17 reported 233 cases from different hospitals on a global scale. Mohammed et al.Citation18 reported 26 cases from the United States and reviewed 97 cases from the literature, and Horn et al.Citation12 described 65 cases from the North American Path Alliance Registry. A major problem in comparative studies is the subdivision of the classical species into a series of molecular siblings, which renders the older literature without sequence data uninterpretable. Despite the current clinical importance of the organism, the phylogenetic relationships among species, varieties and geographical groups in Fusarium are currently elusive. Hence, the re-interpretation of these data in the light of modern molecular phylogeny is compulsory.
Molecular phylogenetic studies have led to the description of many Fusarium species with clinical relevance. These include members of the F. solani species complex, namely, F. falciforme, F. keratoplasticum, F. lichenicola, F. petroliphilum, F. pseudensiforme and F. solani (FSSC5), which is also known as Fusisporium solani and Fusarium haplotype ‘6’. The F. oxysporum species complex (FOSC) contains three lineages, which are involved in fusariosis and still have not been formally introduced as taxonomic species. The F. fujikuroi species complex includes F. acutatum, F. ananatum, F. anthophilum, F. andiyazi, F. fujikuroi s.s., F. globosum, F. guttiforme, F. musae, F. napiforme, F. nygamai, F. verticillioides, F. proliferatum, F. ramigenum, F. sacchari, F. subglutinans, F. temperatum and F. thapsinum. Although rare, species of other Fusarium lineages are emerging as potential opportunistic pathogens, for example, in the F. incarnatum-equiseti species complex (FIESC; F. incarnatum and F. equiseti), the F. dimerum species complex (F. dimerum, F. delphiniodes and F. penzigii), the F. chlamydosporum species complex, the F. sambucinum species complex (F. armeniacum, F. brachygibbosum, F. langsethiae and F. sporotrichioides) and the F. tricictum species complex (F. acuminatum and F. flocciferum).Citation1
Over the past decade, the number of cases of fusariosis has increased worldwide, but there are only a few reports describing the molecular epidemiology; therefore, the aim of the present study is to introduce a hypothetical system that permits the interpretation and use of at least a part of the literature where sequence data are lacking. Pre-molecular publications, which include interpretable case reports and geographical information, were collected. Subsequently, available Fusarium strains that were collected worldwide and deposited during the past century in the CBS-KNAW, Fungal Biodiversity Centre, culture collection Utrecht, The Netherlands, were sequenced and re-identified with current diagnostic technology, which enables the phylogenetic analysis of the human–pathogenic Fusarium species. These data were then compared with published materials and their distribution with the assumption that their distributions in each region had remained unaltered.
MATERIALS AND METHODS
Fungal strains
A total of 127 strains collected from clinical samples (n=74; 58.3%; collected between 1978 and 2015) and strains collected from the environment (n=53; 41.7%; collected between 1929 and 2015) were analyzed. All of the strains were maintained under the name ‘Fusarium’ in the reference collection of CBS-KNAW, Utrecht, the Netherlands. The data regarding geographic origins and sources of isolation are listed in . All of the available type strains were included. Stock cultures were maintained on slants of 2% malt extract agar at 24 °C. The strains were assigned to a clinical subgroup and an environment subgroup.
Table 1 Isolates of Fusarium included in this study that were used for the sequence analysis and amplified fragment length polymorphism analysis, except for six RPB2 and TEF1, which were retrieved from GenBank
DNA extraction
DNA was extracted following the Quick Cetyl trimethylammonium bromide (CTAB) protocol. A total of 1–10 mm3 fungal material was transferred to 2- mL screw-capped tubes prefilled with 490 μL 2 × CTAB buffer and 6–10 acid-washed glass beads. A total of 10 μL of proteinase K was added and mixed thoroughly on a MoBio vortex (MO BIO Laboratories, Inc., Carlsbad, CA, USA) for 10 min. Then, 500 μL of chloroform:isoamylalcohol (24:1) was added and shaken for 2 min after incubation for 60 min at 60 °C. The tubes were centrifuged for 10 min at 14 000 rpm, and the supernatant was collected in a new Eppendorf tube. To ~400 μL of the DNA sample, 2/3 vol (~270 μL) of ice-cold isopropanol was added and centrifuged again at 14 000 rpm for 10 min, and the upper layer was dissolved in 1 mL ice-cold 70% ethanol. The tubes were centrifuged again at 14 000 rpm for 2 min, air-dried and resuspended in 50 μL TE buffer. The quality of the genomic DNA was verified by running 2–3 μL on a 0.8% agarose gel. Then, the DNA was quantified with a NanoDrop 2000 spectrophotometer (Thermo Fisher, Wilmington, DE, USA), and the samples were stored at −20 °C until ready for analysis.
DNA amplification and sequencing
The following two gene regions were amplified directly from the genomic DNA: the second largest subunit of RNA polymerase (RPB2; Reeb et al.Citation19) and the translation elongation factor-1α (TEF1α; O'Donnell et al.Citation20) were amplified and sequenced following the methods published by Saleh et al.Citation16 The PCR reactions were performed in a volume of 12.5 μL containing 1.25 μL of 10 × PCR buffer, 7.5 μL of water, 0.5 μL of dNTP mix (2.5 mM), 0.25 μL of each primer (10 pmol), 0.05 μL of Taq polymerase (5 U/μL), 0.7 μL of dimethylsulphoxide and 1 μL of template DNA (100 ng/μL). The amplification was performed with the ABI Prism 2720 thermal cycler (Applied Biosystems, Foster City, CA, USA). The cycling conditions included 1 cycle of 5 min at 94 °C, 10 cycles of 45 s at 94 °C, 45 s at 55 °C and 1.5 min at 72 °C, 30 cycles of 45 s at 94 °C, 45 s at 52 °C and 1.30 min at 72 °C, a post elongation step of 6 min at 72 °C for TEF1 (EF1, EF2) and a pre-denaturation for 3 min at 95 °C, 5 cycles of 45 s at 95 °C, 45 s at 58 °C and 2 min at 72 °C, 5 cycles of 45 s at 95 °C, 45 s at 56 °C and 2 min at 72 °C, 30 cycles of 45 s at 95 °C, 45 s at 52 °C and 2 min at 72 °C, and a post elongation step of 8 min at 72 °C for RPB2 (5F2 and 7cr). The PCR products were visualized by electrophoresis on 1% (w/v) agarose gels. The sequencing PCR was performed as follows: 1 min at 95 °C followed by 30 cycles consisting of 10 s at 95 °C, 5 s at 50 °C and 2 min 60 °C. The reactions were purified with Sephadex G-50 fine (GE Healthcare Bio-Sciences, Uppsala, Sweden), and the sequencing was conducted on an ABI 3730xL automatic sequencer (Applied Biosystems) with a BigDye v3.1 terminator cycle sequencing kit (Applied Biosystems).
Identification
The strains were identified by BLAST in GenBank, Fusarium MLST (http://www.cbs.knaw.nl/fusarium/)Citation20 and the FUSARIUM-ID (http://isolate.fusariumdb.org/)Citation21 databases. In addition, the phylogenetic placements with species/haplotypes within species complexes were verified with available databases that are specific for Fusarium.
Phylogenetic analyses
Sequences of TEF1 and RPB2 were undertaken to extend the genetic characterization of 127 isolates of Fusarium species previously characterized in terms of morphological characteristics. The sequences were edited using SeqMan in the Lasergene package (DNAstar, Madison, WI, USA). A phylogenetic approach was used to investigate the relationship between 65 strains of Fusarium species including type and reference strains. The sequences were aligned using MAFFT v. 7.127 (http://mafft.cbrc.jp) followed by manual adjustments with MEGA v. 6.2.Citation22 A combined alignment was constructed for RPB2 and TEF1 for both the reference and test strains. The best-fit model of evolution was determined by MEGA v. 6.2.Citation22 A bootstrapped maximum-likelihood analysis was performed using RAxMLVI-HPC v. 7.0.3Citation23 as implemented on the Cipres portal (http://www.phylo.org/),Citation24 with non-parametric bootstrapping using 1000 replicates. Detailed analyses of medically important strains were compared in relation with their clinical cases. For instance, F. solani actually represents a complex (that is, the F. solani species complex).
AFLP
The Fusarium strains were subjected to amplified fragment length polymorphism (AFLP) genotyping using a previously described method.Citation25 However, for the amplification of the DNA fragments, the selective residues (underlined) of the HpyCH4IV-primer (5′-GAT GAG TCC TGA CTA ATG AG-3′) and MseI-primer (5′-Flu-GTA GAC TGC GTA CCC GTAC-3′; MseI-C-selective primer) were replaced. The amplicons were diluted 20 × with double-distilled H2O (ddH2O); 1 μL of the diluted amplicon was then added to a mixture of 8.9 μL ddH2O and 0.1 μL LIZ600 (Applied Biosystems) followed by a heating step for 1 min at 100 °C and cooling to 4 °C. The AFLP fragment analysis was conducted using an ABI3500xL Genetic Analyzer (Applied Biosystems) according to the manufacturer’s instructions. The raw data were then inspected visually after importation into BioNumerics v7.5 (Applied Maths, Sint Martens-Latem, Belgium) and analyzed by an Unweighted Pair Group Method with Arithmetic Mean clustering using the Pearson correlation coefficient. The analysis was restricted to DNA fragments in the range of 40–400 bp. The final AFLP dendrograms were based on the combination of sequencing and the AFLP data of both dendrograms.
Meta-analysis
The authors analyzed the existing medical literature on human cases of fusariosis from 1958 until December 2015. The authors conducted a systematic literature search using PubMed, and the terms ‘Fusarium’ and ‘fusariosis’ were used for the search and both were also used as MeSH words and free words. Studies were only included that reported data for the individual cases because data provided in aggregate often lacked specific information for individual cases. Only cases with either histologically or culturally proven Fusarium infection were included. A total of 388 case reports in ~265 published studies were collected on a worldwide basis. The numbers are approximate because some cases have been used in repeated publications. Only cases with either histologically or culturally proven Fusarium infection were included (Supplementary Reference S1).
RESULTS
Types of articles
A total of 388 cases of fusariosis from 1958 until December 2015 were used in the literature data analysis. This included articles that were mostly single case reports, two patient cases and a series of cases of fusariosis. The reported cases of fusariosis were identified from all over the world, and particularly from tropical and subtropical countries with a large agrarian population such as Brazil, China, Colombia, India and Mexico. The other areas with frequent fusariosis were Australia, South Africa, Turkey and the Americas. Fusarium infections have also been reported from different countries in eastern and western Europe.
Patient characteristics
An overview of the cases of fusariosis published in the medical literature, which includes the great majority of cases published to date, is provided in . The majority of patients were male (n=253; 65.2%; mean 41 years; range three months–83 years). Over a third of the patients (n=143; 36.9%) had various underlying conditions at the time when the Fusarium infection was diagnosed. Causes of immunosuppression were hematological diseases and hematologic malignancies (n=122; 31.4%) and cancer of the solid organs (n=17; 4.8%). Other causes of immunosuppression were medication (n=140; 36%), which included antibiotic (n=34; 8.8%) and steroid treatment (n=10; 2.6%). Pathogen introduction was ranked as trauma (n=18; 4.6%), indwelling catheters (n=2; 0.5%), nasogastric tubes (n=2; 0.5%) and dialysis (n=3; 0.77%). No metabolic disorders, such as diabetes, were recorded in association with infection.
Table 2 Characteristics of 388 patients with fusariosis and literature cases from 1958 until 2015
Type of infections
Infections due to Fusarium were predominantly found to be superficial and subcutaneous (n=174; 44.8%), occurring on the skin (n=62; 16%), eyes (n=66; 17%) and nails (n=25; 6.4%). Deep infections involved bone, joint and lung (n=4; 1%), heart (n=3; 0.77%), and peritoneum (n=2; 0.5%). The sum of the invasive and disseminated cases was n=109 (28%), some of which were associated with fungemia (n= 25; 6.4%) or disseminated disease with brain abscesses (n=4; 1%; ).
Treatment
An overview of the reported treatment of the cases of fusariosis is shown in . The most widely used antifungal agent was amphotericin B deoxycholate (n=198; 51%), followed by liposomal amphotericin B (n=45; 11.6%), voriconazole (n=42; 10.8%), 5-flucytosine (n=30; 7.7%), itraconazole (n=26; 6.7%), fluconazole (n=25; 6.4%) and ketoconazole (n=19; 4.9%).
Table 3 Treatment administered to 388 patients with fusariosis
The antifungal combinations used in treating fusariosis were given either as a two- or a three-drug combination. The most frequently used combination of two drugs was amphotericin B with voriconazole (n=24; 6%), followed by amphotericin B with 5-flucytosine (n=20; 5%), amphotericin B with ketoconazole (n=4; 1%) and amphotericin B with fluconazole (n=4; 1%). Other combinations were used in one or two cases. Triple combinations were used in 14 cases (n=14; 3.6%). In addition, surgery with antifungal treatment was used in 80 cases (20.6%). In addition to antifungal therapy and surgery, granulocyte transfusions or granulocyte–colony-stimulating factor transfusions were also used. Only seven isolates were associated with cases where no treatment was reported (). It was not possible to look at the changes in treatment over time, although the authors assume that azole treatments have increased while AmB has declined. With the current guidelines, liposomal amphotericin B (n=45; 11.6%) and voriconazole (n=42; 10.8%) are very similar according to the data from the reported cases.
Genetic analysis
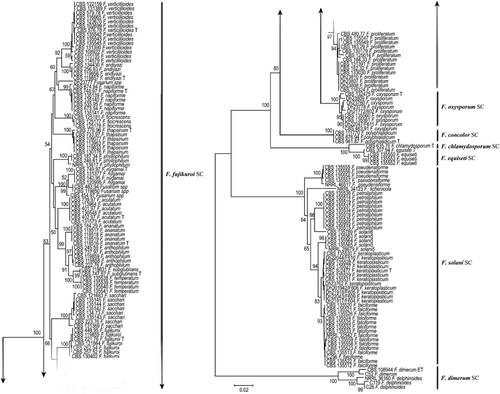
A total of 127 Fusarium strains deposited in the CBS-KNAW collection were partially sequenced for RPB2 and TEF1. The resulting two phylogenies yielded almost identical topologies with similar resolution. Almost all of the strains of known species in all complexes of Fusarium formed independent clades in each tree. A concatenated tree (), including all major human–pathogenic complexes of Fusarium, was based on 146 selected sequences. The lengths of the generated sequence data were 795 and 507 bp for RPB2 and TEF1, respectively. Of the 1302 nucleotides sequenced, 720 (55.1%) were constant, 551 (42.2%) were parsimony informative and 576 (44.1%) were variably and parsimony non-informative using MEGA v. 6.2.Citation22 The combined tree was subdivided into several species complexes with high bootstrap values (). Seven clades represented human opportunists within the F. solani species complex. Thirteen groups represented opportunistic species in the F. fujikuroi species complex with smaller human-associated clusters in the FOSC and to a lesser extent in the F. chlamydosporum, F. polyphialidicum (syn. F. concolor), F. dimerum and F. incarnatum species complexes. Strains CBS 454.97, CBS 483.94 and CBS 119850 were identified morphologically as F. napiforme but formed a separate cluster that was different from the three strains including the type strain of F. napiforme ().
Figure 1 A phylogenetic tree resulting from the RAxML analysis for the RPB2 and TEF1 genes. The total alignment length is 1302 bp. A maximum-likelihood analysis was performed using RAxML with non-parametric bootstrapping using 1000 replicates. The numbers above the branches are bootstrap support values ≥0.70. The outgroup was the epitype (ET) strain of F. dimerum CBS 108944.
The AFLP profiles contained ~50−60 fragments in the range of 40−400 bp. The AFLP dendrogram comprised seven main clusters at the species complex level and additional subgroups within the main species clusters revealed genetic diversity within each species complex (). However, the profiles did not significantly vary between the F. solani species complexes, such as F. falciforme, F. keratoplasticum, F. lichenicola F. petroliphilum and F. pseudensiforme, whereas there was significant AFLP variation between isolates within the F. fujikuroi species complex with separate profiles for each species and within other species complexes of F. chlamydosporum, F. concolor, F. dimerum, F. incarnatum-equiseti and F. oxysporum.
Figure 2 Clustering of the amplified fragment length polymorphism banding pattern of Fusarium spp. combined with a sequence analysis of RPB2 and TEF1 constructed by Bionumerics v7.5 (Applied Maths). The dendrogram was generated using the Unweighted Pair Group Method with Arithmetic Mean algorithm.

When comparing the AFLP clusters with the distribution of DNA sequence lineages, the groups were largely concordant. Groups 1−7 matched with previous identifications using RPB2 and TEF1 sequences. The Fusarium concolor species complex had one clinical subgroup, the F. dimerum species complex had two and the Fusarium fujikuroi species complex consisted of 16 clinical subgroups (15 named subgroups and 1 unnamed molecular lineage). The FIESC had a single clinical group, the FOSC was divided into two subgroups and the F. solani species complex comprised six named and one unnamed subgroup. The AFLP clusters and subclusters were almost identical to the sequencing identifications except for few strains within the F. solani species complex (). The AFLP clusters were based on sequencing and the AFLP data generated from combinations of both dendrograms. In this study, similar clinical presentations of fusariosis were observed among the different AFLP species/genotypes, and there seems to be no relation between species and clinical presentation.
DISCUSSION
The molecular epidemiology of Fusarium was investigated based on the genetic data generated for the RPB2 and TEF1 sequence analysis supplemented with the AFLP fingerprint data. The current study provides information on the locality, sources, species and geographic distribution in countries in which these fungi are common such as Brazil, China, Colombia, India, Mexico and the USA. One hundred and thirty Fusarium isolates, including eight type strains deposited at the CBS-KNAW collection, were included. Overall, there was a good correlation between sequence analysis and AFLP genotypes. To analyze the genetic relationships among the AFLP and sequencing genotypes, we used phylogenetic algorithms, which are commonly used to detect evidence of population subdivision and differentiation.Citation26
The second largest subunit of RNA polymerase (RPB2) and TEF1 showed high resolution with high species-level resolution, distinguishing 29 Fusarium species including the most common clinically important Fusarium species F. falciforme, F. keratoplasticum, F. oxysporum, F. petroliphilum, F. proliferatum and F. verticillioides. Furthermore, these two loci were able to distinguish between other rare Fusarium species that cause a variety of infections ().
Although human opportunists were highlighted in many studies focusing on specific regions of the world and specific types of infections,Citation10, Citation11, Citation12, Citation13, Citation27, Citation28, Citation29, Citation30, Citation31, Citation32, Citation33, Citation34 the 127 Fusarium strains from the current study were collected from 26 countries in six continents and included clinical and environmental strains and isolates from cold blooded animals. Of these, Australia, Brazil, India, Mexico and the USA were among the top 10 countries with the highest Fusarium infections based on clinical isolates in the CBS collection. Not surprisingly, 75 of the 127 patients from this study acquired their infection in one of these countries.
Previously, the majority of the clinically relevant Fusarium species were classified as two species complexes that in the past were referred to as a single species, F. oxysporum and F. solani.Citation35 Approximately 80% of human infections are caused by members of both species complexes,Citation36 but a significant share of infections is caused by the following novel species complex members: F. dimerum, F. fujikuroi and F. incarnatum-equiseti. Within the F. solani complex, there are six recognized species and one unnamed lineage (FSSC6) clinically involved in fusariosis (). Of these species, F. falciforme (n=14/127 cases; 11%) was the dominant species in our study and mainly isolated from keratitis cases in Brazil, India and Mexico. Recently, Hassan et al.Citation13 showed that the majority of keratitis cases (n=46/65 cases; 70.7%) were F. falciforme. This species is emerging as one of the most virulent Fusarium species associated with fusariosis and keratitis.Citation15, Citation36, Citation37
In the 2005–2006 mycotic keratitis outbreaks in Southeast Asia and North America that were associated with a contact lens cleaning solution, F. petroliphilum and F. keratoplasticum were the most common species,Citation36 which is consistent with the current study. The AFLP genotypic variability was higher in the environmental species than in the clinical species. A potential explanation is that not all environmental genotypes are sufficiently adapted to the host tissue and are not selected or perhaps a sampling effect is involved. Zhang et al.Citation35 studied the F. solani species complex, specifically those species that cause infections in humans and plants, and concluded that clinical isolates often shared multi-locus haplotypes with isolates from different environmental sources, including hospital locations. An increase of fusariosis among immunosuppressed patients was noted in the bone marrow transplant unit and among patients with superficial infections in a hospital in Rio de Janeiro, Brazil.Citation38 These authors concluded that this increase might be due to airborne conidia circulating in this geographical region. Short et al.Citation36 concluded that there is no evidence that clinical isolates differ from those collected from other sources.
The large diversity of the FOSC is not completely resolved, and it is not yet known whether the species have one or several phylogenetic origins or whether a single species or a species complex is concerned. From a traditional taxonomic point of view, F. oxysporum isolates are differentiated from each other based on the pathogenicity as formae speciales, but this has been shown to be an unreliable approach.Citation8 In addition, the species delimitation was for the FOSC, and at least 26 sequence types within the complex were involved in human infections.Citation39 Our FOSC clinical isolates were distributed throughout the complex, although some clustering was found in the clade marked ‘sequence type 33’, which is based on TEF1 alone, and this sequence type is considered the most common clade that contains clinical F. oxysporum strains. The remaining species complexes of F. chlamydosporum, F. concolor, F. dimerum and F. incarnatum-equiseti form separate clusters in the highly resolved sequence-based maximum likelihood tree ().
The FIESC compromises 28 phylogenetically distinct lineages,Citation34 and only 2 are named and mainly involved in human infections (F. incarnatum and F. equiseti).Citation40 Although several members of the FIESC were included in the CDC Fusarium keratitis outbreak investigations, these species have not yet been observed to occur in epidemics or cause outbreaks. Concerning geography, 51 clinical isolates were recovered from the United States, and this revealed that phylogenetically diverse human opportunists are well represented in North America.Citation40 In our study, three clinical F. equiseti strains originated from Mexico, and this might suggest that species of this complex are common in this region. The virulence of members of the FIESC has been ascribed to their production of type A and B trichothecene mycotoxins.Citation39
F. dimerum and F. delphinoides belong to the F. dimerum species complex, and both were isolated from superficial and disseminated infections.Citation15 In our data set, a supported clade of FDSC matching with the AFLP data mainly contained strains from India, and this might suggest a regional prevalence. F. chlamydosporum was reported in disseminated infections in patients with aplastic anemia and lymphocytic lymphoma from the United StatesCitation41, Citation42 CBS 111770 (F. concolor) is the only clinical strain in the F. concolor species complex, and it was reported in a keratitis case from Spain.Citation43
By comparing AFLP and MLST data, F. falciforme and F. keratoplasticum appear to be widely distributed, at least in Mexico, North America, Europe and India, with dominancy in superficial infections, including keratitis and onychomycosis. F. petroliphilum is the second most diverse species and is also frequently involved in disseminated infections. F. solani sensu stricto, ‘5’, which was recently described as Fusisporium (Fusarium) solani (FSSC5),Citation7 contains strains such as CBS 135559, CBS 135564 and CBS 135565, which originate from Mexico, and shows significant occurrence in keratitis cases. This species was also recently reported in Asia (India and Qatar).Citation13, Citation16 Given the large distances of identical strains occurring in many different countries, airborne distribution seems likely. However, the presence of F. incarnatum, F. equiseti and F. chlamydosporum in clinical samples from various infections in North America remains puzzling but can perhaps be explained by sampling effects.
As previously noted,Citation44 the F. fujikuroi complex contains the highest number of species. In our study, 15 supported clades were recognized in all of the molecular analyses ( and ). Nearly all of the clades have various geographic distributions. Within the F. fujikuroi species complex, F. proliferatum and F. verticillioides were the dominant clinically relevant species, having a global distribution and dominating in disseminated infections. F. sacchari is the second most prevalent species and was often isolated from keratitis restricted to India. Although F. nygamai and F. napiforme are the most multidrug-resistant species within the F. fujikuroi complex,Citation45 their presence in human infections is rare. F. acutatum was reported from nail infections in four cases in Qatar, showed a low degree of variability and has been suggested to be clonal.Citation16 These results emphasize that F. acutatum is an emerging human opportunist, which thus far was only detected in Asia. Sequence analysis of RPB2 and TEF1, and AFLP showed that the strains CBS 119850, CBS 483.94 and CBS 454.97 were nested within the F. fujikuroi complex and close to F. nygamai and F. andiyazi, forming a well-supported monophyletic branch suggestive of a novel species.
Deep fusariosis is rare in healthy individuals; a single brain infection has been reported.Citation46 Local infections may occur after a direct inoculation or tissue breakdown by trauma or the entrance of foreign bodies. The treatment of superficial infections is usually successful and requires surgery, the removal of the foreign body and antifungal therapy. The most important risk factors for severe fusariosis are prolonged neutropenia and T-cell immunodeficiency in patients suffering from a hematologic malignancy. Fusarium infections in the majority of these cases were due to neutropenia. Furthermore, in solid organ transplant recipients and cancer patients with neutropenia, infections due to Fusarium spp. increased and led to disseminated infection. Patients develop painful skin lesions, which vary from papules to nodules with or without central necrosis.Citation47 In the majority of disseminated infections, secondary skin lesions led to a diagnosis in >50% of the patients and preceded fungemia by ~5 days.Citation48 In contrast to aspergillosis, fusariosis frequently shows positive blood cultures because Fusarium conidia are hydrophilic and allow dissemination.Citation47 Comparing fusariosis with mucormycosis,Citation49 solid tumors and diabetes do not seem to be important risk factors. Only 17 (4.8%) cases were found in patients with solid tumors, and seven infections were reported in patients with diabetes mellitus. No underlying conditions were observed in 20 (5%) of the cases.
Fusarium treatment depends on the site of infection. Surgery with antifungals was used in 80 cases (20.6%). Disseminated fusariosis in immunocompromised patients is usually treated with amphotericin B and voriconazole as the first-line therapy, which is suggested by recent guidelines.Citation50 In our literature review, most antifungal therapy was amphotericin B deoxycholate, followed by liposomal amphotericin B and voriconazole. The most commonly used combination is amphotericin B/voriconazole followed by amphotericin B/5-flucytosine. Triple combinations were used in 14 cases with different antifungals.
The major findings of the present study include the following: (i) human-associated fusaria were nested within seven species complexes (that is, F. chlamydosporum, F. concolor, F. dimerum, F. fujikuroi, F. incarnatum-equiseti, F. oxysporum and F. solani), (ii) the three most common species presented in both the clinical and environmental groups are F. falciforme and F. keratoplasticum (members of F. solani species complex) followed by F. oxysporum (FOSC), (iii) most of the reported Fusarium species in this study were shared among the patients and the environment, and this might be due to the colonization of some patients with Fusarium isolates from the environment; hence, there is genetic similarity between the clinical and environmental isolates of the same Fusarium species, and (iv) the species distributions show some evidence of geographical clustering among some of the species studied, although the present study is limited by an over-representation of isolates from Mexico and India.
Supplementary Methods
Download MS Word (58.5 KB)This study was partially funded by the Ministry of Health, Oman (Formal Agreement NO 28/2014). Abdullah MS Al-Hatmi received a PhD scholarship from the Ministry of Health, Oman.
Supplementary information accompanies the paper on Emerging Microbes and Infections's website (http://www.nature.com/emi).
- Al-HatmiAM,MeisJF,de HoogGS.Fusarium: molecular diversity and intrinsic drug resistance.PLoS Pathog2016; 12:e1005464.
- NucciM,VaronAG,GarnicaMet al.Increased incidence of invasive fusariosis with cutaneous portal of entry, Brazil.Emerg Infect Dis2013; 19:1567–1572.
- MukherjeePK,ChandraJ,YuCet al.Characterization of Fusarium keratitis outbreak isolates: contribution of biofilms to antimicrobial resistance and pathogenesis.Invest Ophthalmol Vis Sci2012; 53:4450–4457.
- ShortDPG,O’DonnellK,ZhangNet al.Widespread occurrence of diverse pathogenic types of the fungus Fusarium in bathroom plumbing drains.J Clin Microbiol2011; 49:4264–4272.
- O’DonnellK,SuttonDA,FothergillAet al.Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex.J Clin Microbiol2008; 4:2477–2490.
- Al-HatmiAMS,MeletiadisJ,Curfs-BreukerIet al.In vitro combinations of natamycin with voriconazole, itraconazole and micafungin against clinical Fusarium isolates causing keratitis.J Antimicrob Chemother2016; 71:953–955.
- SchroersH-J,SamuelsGJ,ZhangNet al.Epitypification of FusisporiumFusariumsolani and its assignment to a common phylogenetic species in the Fusarium solani species complex.Mycologia2016; 108:806–819.
- GarnicaM,NucciM.Epidemiology of fusariosis.Curr Fungal Infect Rep2013; 7:301–305.
- O’DonnellK,SuttonDA,RinaldiMGet al.Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin.J Clin Microbiol2004; 42:5109–5120.
- NucciM,GarnicaM,GloriaABet al.Invasive fungal diseases in haematopoietic cell transplant recipients and in patients with acute myeloid leukaemia or myelodysplasia in Brazil.Clin Microbiol Infect2013; 19:745–751.
- NucciM,AnaissieE.Cutaneous infection by Fusarium species in healthy and immunocompromised hosts: implications for diagnosis and management.Clin Infect Dis2002; 35:909–920.
- HornDL,FreifeldAG,SchusterMGet al.Treatment and outcomes of invasive fusariosis: review of 65 cases from the PATH Alliance registry.Mycoses2014; 57:652–658.
- HassanAS,Al-HatmiAMS,ShobanaCSet al.Antifungal susceptibility and phylogeny of opportunistic members of Fusarium species causing human keratomycosis in India.Med Mycol2016; 54:287–294.
- Daylan CiloB,Al-HatmiAM,SeyedmousaviSet al.Emergence of fusariosis in a university hospital in Turkey during a 20-year period.Eur J Clin Microbiol Infect Dis2015; 34:1683–1691.
- Guevara-SuarezM,Cano-LiraJF,de GarciaMCCet al.Genotyping of Fusarium isolates from onychomycosis in Colomobia: detection of two new species within the Fusarium solani species complex and in vitro antifungal susceptibility testing.Mycopathologia2016; 181:165–174.
- SalahH,Al-HatmiAMS,TheelenBet al.Phylogenetic diversity of human pathogenic Fusarium and emergence of uncommon virulent species.J Infect2015; 71:658–666.
- NucciM,MarrKA,VehreschildMJet al.Improvement in the outcome of invasive fusariosis in the last decade.Clin Microbiol Infect2014; 20:580–585.
- MuhammedM,AnagnostouT,DesalermosAet al.Fusarium infection: report of 26 cases and review of 97 cases from the literature.Medicine2013; 92:305–316.
- ReebV,LutzoniF,RouxC.Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory.Mol Phylogenet Evol2004; 32:1036–1060.
- O'DonnellK,SuttonDA,RinaldiMGet al.Internet-accessible DNA sequence database for identifying fusaria from human and animal infections.J Clin Microbiol2010; 48:3708–3718.
- GeiserDM,Jimenez-GascoMD,KangSCet al.FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium.Eur J Plant Pathol2004; 110:473–479.
- TamuraK,StecherG,PetersonDet al.MEGA6: Molecular Evolutionary Genetics Analysis version 6.0.Mol Biol Evol2013; 30:2725–2729.
- StamatakisA.RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models.Bioinformatics2006; 22:2688–2690.
- StamatakisA,HooverP,RougemontJ.A rapid bootstrap algorithm for the RAxML Web servers.Syst Biol2008; 57:758–771.
- Al-HatmiAMS,MirabolfathyM,HagenFet al.DNA barcoding, MALDI-TOF and AFLP data support Fusarium ficicrescens as a distinct species within the F. fujikuroi species complex.Fungal Biol2016; 120:265–278.
- Al-HatmiAMS,Van Den EndeAH,StielowJBet al.Two novel DNA barcodes for species recognition of opportunistic pathogens in Fusarium.Fungal Biol2015; 120:231–245.
- MigheliQ,BalmasV,HarakHet al.Molecular phylogenetic diversity of dermatologic and other human pathogenic fusarial isolates from hospitals in northern and central Italy.J Clin Microbiol2010; 48:1076–1084.
- NinetB,JanI,BontemsOet al.Molecular identification of Fusarium species in onychomycoses.Dermatology2005; 210:21–25.
- LópezNC,CasasC,SopoLet al.Fusarium species detected in onychomycosis in Colombia.Mycoses2008; 52:350–356.
- van DiepeningenAD,FengP,AhmedSet al.Spectrum of Fusarium infections in tropical dermatology evidenced by multilocus sequencing typing diagnostics.Mycoses2015; 58:48–57.
- HomaM,ShobanaCS,SinghYRet al.Fusarium keratitis in South India: causative agents, their antifungal susceptibilities and a rapid identification method for the Fusarium solani species complex.Mycoses2013; 56:501–511.
- Vanzzini ZagoV,Manzano-GayossoP,Hernandez-HernandezFet al.Mycotic keratitis in an eye care hospital in Mexico City.Rev Iberoam Micol2010; 27:57–61.
- LortholaryO,ObengaG,BiswasPet al.International retrospective analysis of 73 cases of invasive fusariosis treated with voriconazole.Antimicrob Agents Chemother2010; 54:4446–4450.
- O’DonnellK,SuttonDA,RinaldiMGet al.Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States.J Clin Microbiol2009; 47:3851–3861.
- ZhangN,O’DonnellK,SuttonDAet al.Members of the Fusarium solani species complex that cause infection in both humans and plants are common in the environment.J Clin Microbiol2006; 44:2186–2190.
- ShortDPG,O’DonnellK,GeiserDM.Clonality recombination, and hybridization in the plumbing-inhabiting human pathogen Fusarium keratoplasticum inferred from multilocus sequence typing.BMC Evol Biol2014; 14:91.
- OechslerRA,YamanakaTM,BispoPJet al.Fusarium keratitis in Brazil: genotyping, in vitro susceptibilities, and clinical outcomes.Clin Ophthalmol2013; 7:1693–1701.
- ScheelC,HurstS,BarreirosGet al.Molecular analyses of Fusarium isolates recovered from a cluster of invasive mold infections in a Brazilian hospital.BMC Infect Dis2013; 13:49.
- O’DonnellK,GueidanC,SinkSet al.A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex.Fungal Genet Biol2009; 46:936–948.
- GuarroJ.Fusariosis, a complex infection caused by a high diversity of fungal species refractory to treatment.Eur J Clin Microbiol Infect Dis2013; 32:1491–1500.
- KiehnTE,NelsonPE,BernardEMet al.Catheter-associated fungemia caused by Fusarium chlamydosporum in a patient with lymphocytic lymphoma.J Clin Microbiol1985; 21:501–504.
- SegalBH,WalshTJ,LiuJMet al.Invasive infection with Fusarium chlamydosporum in a patient with aplastic anemia.J Clin Microbiol1998; 36:1772–1776.
- GuarroJ,RubioC,GenéJet al.Case of keratitis caused by an uncommon Fusarium species.J Clin Microbiol2003; 41:5823–5826.
- Al-HatmiAM,NormandAC,van DiepeningenADet al.Rapid species-level identification of opportunists in the Fusarium fujikuroi species complex using MALDI-TOF mass spectrometry.Future Microbiol2015; 10:1939–1952.
- Al-HatmiAMS,van DiepeningenA,Curfs-BreukerIet al.Specific antifungal susceptibility profiles of opportunists in the Fusarium fujikuroi complex.J Antimicrob Chemother2015; 70:1068–1071.
- GarciaRR,MinZ,NarasimhanSet al.Fusarium brain abscess: case report and literature review.Mycoses2015; 58:22–26.
- NucciM,AnaissieE.Fusarium infections in immunocompromised patients.Clin Microbiol Rev2007; 20:697–704.
- DignaniMC,AnaissieEJ.Human fusariosis.Clin Microbiol Infect2004; 10:67–75.
- RombauxP,EloyP,BertrandBet al.Lethal disseminated Fusarium infection with sinus involvement in the immunocompromised host: case report and review of the literature.Rhinology1996; 34:237–241.
- TortoranoAM,RichardsonM,RoilidesEet al.ESCMID and ECMM joint guidelines on diagnosis and management of hyalohyphomycosis: Fusarium spp., Scedosporium spp. and others.Clin Microbiol Infect2014; 20(Suppl 3):27–46.
