Abstract
Surveillance for avian influenza viruses (AIVs) in poultry and environmental samples was conducted in four live-bird markets in Cambodia from January through November 2013. Through real-time RT-PCR testing, AIVs were detected in 45% of 1048 samples collected throughout the year. Detection rates ranged from 32% and 18% in duck and chicken swabs, respectively, to 75% in carcass wash water samples. Influenza A/H5N1 virus was detected in 79% of samples positive for influenza A virus and 35% of all samples collected. Sequence analysis of full-length haemagglutinin (HA) and neuraminidase (NA) genes from A/H5N1 viruses, and full-genome analysis of six representative isolates, revealed that the clade 1.1.2 reassortant virus associated with Cambodian human cases during 2013 was the only A/H5N1 virus detected during the year. However, multiplex reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of HA and NA genes revealed co-circulation of at least nine low pathogenic AIVs from HA1, HA2, HA3, HA4, HA6, HA7, HA9, HA10 and HA11 subtypes. Four repeated serological surveys were conducted throughout the year in a cohort of 125 poultry workers. Serological testing found an overall prevalence of 4.5% and 1.8% for antibodies to A/H5N1 and A/H9N2, respectively. Seroconversion rates of 3.7 and 0.9 cases per 1000 person-months participation were detected for A/H5N1 and A/H9N2, respectively. Peak AIV circulation was associated with the Lunar New Year festival. Knowledge of periods of increased circulation of avian influenza in markets should inform intervention measures such as market cleaning and closures to reduce risk of human infections and emergence of novel AIVs.
Emerging Microbes & Infections (2016) 5, e70; doi:10.1038/emi.2016.69; published online 20 July 2016
INTRODUCTION
Avian influenza viruses (AIVs) naturally infect the gastrointestinal tracts of wild birds from the Orders Anseriformes (ducks, geese and swans) and Charadriformes (waders and gulls). Within these hosts, 16 haemagglutinin (HA) and nine neuraminidase (NA) surface glycoprotein types have been described. The HA and NA types carried by the virus are used for classification and can be found in various combinations, such as A/H5N1 and A/H3N2. Wild birds have been implicated in the global dissemination of AIVs and, in some instances, the introduction of AIVs into domestic poultry populations.Citation1 Although low pathogenic avian influenza (LPAI) viruses (which have limited impact on poultry mortalities) constitute the majority of these viruses, highly pathogenic avian influenza (HPAI) viruses such as A/H5N1 can result in mortalities approaching 100% when introduced into domestic flocks.Citation2
Influenza A viruses, which constitute all of the known AIVs, have a segmented genome with eight separate RNA strands enclosed in the virus. When an animal is co-infected with two different influenza viruses, reassortment of these RNA segments can occur, resulting in a new virus with characteristics distinct from the parent viruses.Citation3 The 2009 emergence and resulting pandemic from A/H1N1pdm09 virus was caused by a virus that underwent multiple reassortment events with strains from pigs, birds and humans before suddenly gaining the ability to efficiently transmit between humans.Citation4 Pandemic events resulting from reassortment of influenza viruses have occurred at least three times in the last 100 years,Citation5, Citation6 resulting in high human morbidity and mortality worldwide. The most severe influenza pandemic in recorded history, the 1918 ‘Spanish flu’, is thought to have arose from an avian influenza strain that directly adapted to the human host.Citation7 Clearly it is imperative that close monitoring of reassortments and mammalian-adapted mutations in avian influenza strains is needed.
Since the emergence of HPAI A/H5N1 in Southern China in 1996 and 1997,Citation8 descendants of this virus have caused considerable economic losses in poultry populations, primarily in east and southeast Asia, but also in Africa and Europe. Influenza A/H5N1 has been identified as a significant threat for pandemic emergence. To date, at least 844 people have been infected worldwide, resulting in 449 deaths (53% case fatality rate). The virus was first detected in Cambodian poultry in early 2004 and the first human cases were detected in 2005.Citation9 As of December 2015, 56 human cases (including 37 deaths) and 43 poultry outbreaks of influenza A/H5N1 have been recorded in the country.Citation10, Citation11
Live-bird markets (LBMs) have been implicated as an important source of human sporadic cases and dissemination of avian influenza since the emergence of influenza A/H5N1.Citation12 Previous studies have established that LBMs serve as hubs for the circulation and persistence of AIVs through the presence of multiple avian species, the constant introduction of immunologically naive hosts and the frequent lack of biosecurity measures.Citation1, Citation13, Citation14 Recent surveillance studies have documented that A/H5N1 and A/H7N9 commonly co-circulate with other subtypes of avian influenza in LBMs,Citation15, Citation16, Citation17 thus increasing the likelihood of reassortment events.
In the present study we initiated poultry and environmental surveillance for A/H5N1 and other AIVs in four LBMs in Cambodia to better ascertain virus circulation in this setting. A longitudinal human serosurvey was also conducted to investigate the risk of human avian influenza infections in exposed LBM workers.
MATERIALS AND METHODS
Animal and environmental samples
Commencing in January 2013, through November 2013, environmental and poultry samples were collected at four LBMs in Cambodia: an LBM in central Phnom Penh (M1), a wholesaling farm/slaughter house in Phnom Penh (M2), an LBM in Kampong Cham province (M3) and an LBM in Takeo province (M4) (Figure ). These LBMs were selected for the study as they represent the largest poultry collection sites in the most densely populated region of the country. Samples were collected from each market every 1–2 weeks (Figure ). The LBMs in Cambodia typically have poor biosecurity, with slaughtering of animals onsite occurring for a range of domestic animals (Figure ). Birds are typically sourced from backyard flocks by middlemen who transport the animals through a convoluted system of semi-commercial farms and stock houses before being transported to the main LBMs.Citation18 Thus, tracking the original source of poultry is usually not possible.
Figure 1 Map showing communes with reported confirmed human A/H5N1 cases during 2006–2014, population density (indicated by the number of villages) and live-bird markets investigated in 2013.
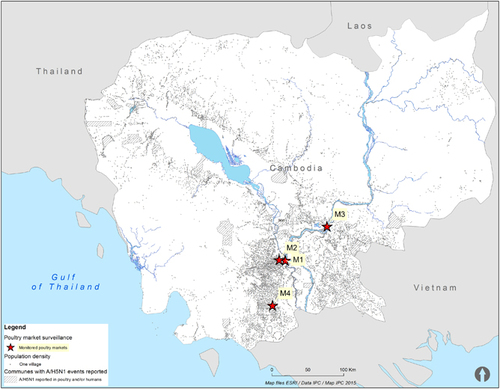
Figure 2 Live-bird markets in Cambodia typically have poor biosecurity, with multiple animal species slaughtered onsite. Animal and environmental sampling in these markets is crucial to monitor virus circulation and the emergence/introduction of new viruses.

During each market investigation, tracheal and cloacal swabs were collected (both samples pooled in one tube of viral transport medium for each animal) from four randomly selected poultry (three ducks and one chicken). Environmental samples were also collected in the same cage/site where the poultry swabs were collected to investigate contamination of the LBMs with AIVs. During each mission, 50 ml of carcass wash water (water used to wash the carcasses once the poultry has been slaughtered and defeathered), 50 ml of poultry drinking water (small bowl of water placed in cages), soil/mud from an area within 50cm around the poultry cages/poultry resting area and samples of discarded feathers were collected. The water, soil/mud and feather samples were processed and nucleic acids extracted following techniques described previously.Citation19, Citation20 Extracts were then tested by quantitative real-time reverse-transcriptase polymerase chain reaction (qRT-PCR) for the detection of M, H5 and N1 genes.
M-gene-positive samples, for which there was sufficient sample and high virus concentration (CT<30; n=78), were inoculated into specific pathogen-free embryonated chicken eggs for virus isolation.Citation21 Isolates were then tested using influenza universal multiplex RT-PCR assays to test for all known subtypes of influenza.Citation22 Universal multiplex RT-PCR typing assays were also applied to the original samples where isolation could not be achieved but discordance between M-gene and H5 qRT-PCR testing suggested the presence of a non-H5 AIV. The amplified RT-PCR products from isolates and original material were submitted to a commercial sequencing facility (Macrogen, Seoul, South Korea) for sequencing by the Sanger method. Full-genome sequences were generated from representative A/H5N1 isolates using methods previously described.Citation9, Citation23
Contiguous sequences were assembled using CLC Workbench (CLC bio) and compared to representative influenza virus sequences downloaded from the NCBI GenBank database. Neighbour-joining trees were constructed with MEGA5Citation24 and bootstrap values were calculated and expressed as a percentage from 1000 replicates.
Human samples
The longitudinal human serological study was approved by the National Ethics Committee for Human Research (approval NO. 267, 24 December 2012). Serum samples were collected, after obtaining informed consent, from LBM workers at the start of the study (January 2013) to form a baseline, and 8 weeks after the three major national festivals shown by previous work to be associated with increased A/H5N1 circulation in markets:Citation25 Lunar New Year, week 6; Khmer New Year, week 15; Pchum Ben, week 40. All adult-age LBM sellers or workers were exhaustively recruited in the four targeted live-poultry markets. The sample size could not be calculated as transmission to humans was unlikely and its probability in Cambodia is unknown. Participants were informed to report any acute febrile, respiratory or digestive signs, and were provided with a toll-free phone number.
Serum samples were tested for avian influenza A/H5N1, A/H9N2 and A/H7N9 antibodies using the haemagglutination inhibition assay (HIA) and microneutralization assay (MN). The HIA and MN testing were performed using influenza A/H5N1 clade 1.1.2 reassortant viruses (A/Cambodia/X0121311/2013 and A/Cambodia/ X0125302/2013), which were isolated from human cases during 2013, and influenza A/H9N2 virus (A/Environment/Cambodia/E265/2013), which was isolated from the LBMs during 2013. The HIA and MN testing for A/H7N9 were performed using the strain A/Anhui/01/2013, supplied through the World Health Organization (WHO) Global Influenza Surveillance and Response System (Dr Sylvie van der Werf, Department of Virology, Institut Pasteur, Paris, France). Exposure to these viruses was considered ‘confirmed’ with a HIA titre of ≥80 and a MN titre of ≥40. A seroconversion was defined as the detection of antibodies equal to or above the thresholds defined above following no detection of antibodies in the serum sample from the previous period.
Laboratory data were entered using an Excel spreadsheet (Microsoft Excel, Microsoft, Redmond, WA, USA). A baseline assessment was documented using point prevalence for influenza antibodies, and incidence rates during follow-up were then computed using data on laboratory-confirmed seroconversions in LBM workers (numerator), and the number of days elapsed during the last serosurvey (denominator). Poisson confidence intervals for the incidence rates were computed using Stata 11 (Stata Corp., College Station, TX, USA) with the function cii for the binomial CI, and the functions cii and the option ‘poisson’ for the poisson CI.
RESULTS
Animal and environmental surveillance of A/H5N1
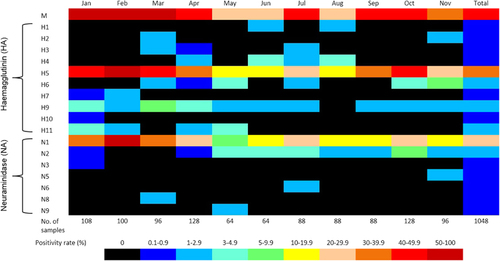
During the study, a total of 1048 samples were collected, with 45% of all samples positive for influenza A RNA by qRT-PCR (Table ). Influenza A viruses were detected in at least one sample during 93% of 120 collection missions, with influenza virus detected in three of the markets (M1, M2 and M3) on all but one sampling mission and detected in 83% of sampling missions from the remaining market (M4). Influenza A (M-gene) ribonucleic acid (RNA) was detected most frequently in carcass wash water samples (75%, n=146), followed by feathers (61%, n=138), poultry drinking water (50%, n=138), soil/mud (48%, n=146), duck swabs (32%, n=358) and chicken swabs (18%, n=122). H5 and N1 genes were detected in 79% (n=372) and 58% (n=270), respectively, of the 468 samples that tested positive for influenza A (M-gene), which accounted for 35% and 26% of all samples collected, respectively. Peak AIV circulation was detected from January through March, with particularly high detection rates during the Lunar New Year festival period (Figure ).
Figure 3 Proportion of samples positive for Influenza A virus (M-gene) for each sampling mission at four live-bird markets in Cambodia, 2013. The major Cambodian festivals are during week 6 (Lunar New Year festival), week 15 (Khmer New Year festival) and week 40 (Pchum Ben festival), and are indicated by vertical blue bars.
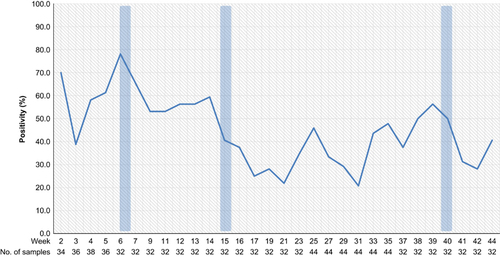
Table 1 Results of qRT-PCR tests on environmental samples and poultry swab specimens collected from four live poultry markets in Cambodia, 2013 (results displayed by sample type and by market)
Influenza A/H5N1 virus was isolated from 71% (55/78) of samples from which H5 was detected and isolation was attempted. Isolation was only attempted on high viral load samples (M-gene qRT-PCR CT value <30) and success rates differed considerably by sample type, with isolation being highest in duck swabs (83%, 19/23), followed by poultry drinking water (80%, 8/10), soil/mud (72%, 13/18), feathers (64%, 7/11), carcass wash water (53%, 8/15) and chicken swabs (0%, 0/1).
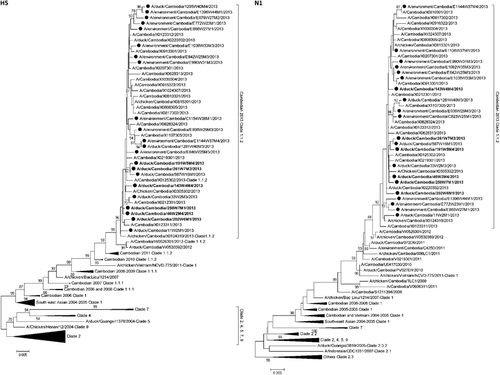
Full-gene sequences were generated for H5 (n=22) and N1 (n=21) for all viruses detected in the study where sufficient virus concentration allowed for successful sequencing (Figure ).Citation10, Citation23 Full-genome sequences (eight fragments) were generated for six influenza A/H5N1 viruses detected during the study (Supplementary Figure S1). For other viruses full-gene sequences could not be generated for all fragments (Supplementary Table S1). All A/H5N1 viruses detected in the study clustered with the clade 1.1.2 reassortant strains associated with human cases and poultry outbreaks of A/H5N1 during 2013.Citation10
Figure 4 Neighbour-joining phylogenetic tree of the HA and NA genes of highly pathogenic avian influenza A/H5N1 viruses detected during live-bird market surveillance in Cambodia. Viruses collected during the present market study are denoted by a black circle. Viruses for which the full genome is available are denoted in bold. Mid-point rooted phylogenetic trees were constructed in MEGA5. Bootstraps >70 generated from 1000 replicates are shown at branch nodes. The scale bar represents the number of nucleotide substitutions per site.
Non-H5 avian influenza detection
A large number and variety of non-H5 avian influenza strains were detected during the study. Full-HA sequences were generated from two H1, two H2, three H3, nine H4, 15 H6, one H7, 27 H9, one H10 and twelve H11 viruses (Supplementary Figure S2). Full NA sequences from twenty-two N2, one N3, one N5, two N6, one N8 and two N9 subtypes were also generated from the same samples (Supplementary Figure S3). However, as many of the environmental samples contained evidence of multiple avian influenza strains, it was difficult to ascertain that HA and NA sequences derived from the same sample actually belonged to the same virus strain. Co-infections between AIVs were not detected in any poultry samples. Many other partial HA and NA sequences were also generated, but not included in these results due to difficulties in determining close phylogenetic relationships from incomplete gene sequences. AIVs were detected throughout the year (Figure ), with a distinct peak of activity during January−March, perhaps mostly due to the increased circulation of A/H5N1 during major Cambodian festivals, as previously reported.Citation25
Longitudinal human serosurvey
Successive serological surveys in the poultry worker cohort provided evidence of seroconversions and some prior exposure (Table ). At baseline sampling (January 2013), 125 participants were enrolled in the study, with one person testing positive to A/H5N1 antibodies and another testing positive to A/H9N2 antibodies. Participant retention was high throughout the study, with 117, 105 and 106 people resampled at the second (March 2013), third (June 2013) and fourth (November 2013) sampling missions, respectively. Seroconversions to A/H5N1 were detected for two participants at the third sampling and two participants at the fourth sampling. Seroconversions to A/H9N2 were detected for one participant at the third sampling. Overall seroprevalence was 4.5% for A/H5N1 and 1.8% for A/H9N2. Rates of seroconversion were 3.7 infections per 1000 person-months for A/H5N1 and 0.9 infections per 1000 person-months for A/H9N2. There was no serological evidence of exposure or molecular detection of A/H7N9 before or during the study.
Table 2 Positive test results in participants and estimated seroprevalence and incidence rate, Cambodian live-bird markets study, 2013
There were no calls to the toll-free number reporting an incident with clinical signs or symptoms compatible with influenza infection.
DISCUSSION
LBMs have been established as important foci for the transmission of AIVs and the potential emergence of reassortant strains.Citation13 The presence of multiple host species and the continued introduction of naive birds create an ideal environment for the persistence and emergence of AIVs. Exposure to live poultry has been associated with fatal human A/H5N1 infections, which for instance led to the government of Hong Kong rapidly closing LBMs in 1997 and slaughtering large numbers of poultry.Citation26 The LBMs surrounding Phnom Penh have been established as foci for poultry movement in the countryCitation18, Citation27 and a previous market surveillance study in 2011 revealed a high rate of influenza A/H5N1 circulation in this setting.Citation25
To our knowledge, AIVs were detected in Cambodian LBMs during 2013 at a higher frequency than any other study published previously,Citation25, Citation28, Citation29, Citation30, Citation31, Citation32, Citation33, Citation34 with 45% of all samples (poultry swab and environmental samples), 32% of individual duck swabs and 18% of chicken swabs positive for influenza A RNA. The majority of the AIVs detected were likely A/H5N1, with 35% of samples positive for H5 qRT-PCR. Although there was clearly high co-circulation of LPAI viruses, much of the difference between the detection rates of M-gene, H5 and N1 was probably due to the differing qRT-PCR assay sensitivities (M>H5>N1). We previously detected high circulation of A/H5N1 in these same markets in 2011.Citation25 However, in 2013 there was a >2.5-fold increase in the frequency with which the virus was detected despite the same sample processing, nucleic acid extraction and RT-qPCR methods being used.
In 2013, Cambodia had the highest confirmed human A/H5N1 caseload per capita in the World. During 2013 alone, 26 human A/H5N1 cases (14 deaths) were detected in Cambodia, a dramatic increase in the 21 total cases that had been detected in the preceding 8 years, 2005–2012. This rise in reported human cases of A/H5N1 coincided with the emergence of a reassortant virus that contained the HA and NA genes from the previously circulating clade 1.1.2 genotype Z virus, and the matrix and internal genes from a clade 2.3.2.1 virus previously circulating in southern Vietnam.Citation10 Sequence and phylogenetic analyses of the HA and NA genes (Figure 4) and the matrix and internal genes (Supplementary Figure S1) from the A/H5N1 market strains revealed that all of the viruses clustered closely with other clade 1.1.2 reassortant strains associated with poultry outbreaks and human cases in 2013.Citation10 Questions remain regarding the causes of the dramatic increase in human cases in 2013 and whether the reassortant strain is more transmissible in poultry, resulting in the intense circulation that we observed in this report. Alternatively, increased surveillance and education of clinicians may have resulted in improved detection of human A/H5N1 cases. Presently we do not know the exact role played by LBMs in the infection of poultry workers and market clients.
Similarly to our previous LBM surveillance in 2011,Citation25 increased circulation of AIVs was detected before and during the major Cambodian festivals (Figure 3). In particular, increased circulation of influenza A viruses was detected during the period between the Lunar New Year and Khmer New Year festivals. A previous study on the poultry trade links in Cambodia established that there was a significant increase in the trade volume of poultry prior to these festivals, with an assumed rise in cross-border poultry trade in response to increased demand.Citation18 The greater volume of poultry trade and the expansion of poultry trading networks during these festival periods present opportunities for virus spread throughout domestic flocks and the introduction of new strains of AIVs. Peaks in AIV levels were also observed during times that were not associated with known festivals (e.g. week 25 and 35). Climatic factors, which were not analysed in this study, are also likely to have an influence on levels of AIV circulation. Knowledge of these periods of intense circulation should inform future control policies such as targeted poultry vaccination, quarantining and improved market cleaning. Such measures have proven effective in reducing avian influenza viral isolation rates in LBMsCitation35, Citation36, Citation37 and may reduce the spread of AIVs back to farms through fomites and personnel.Citation38
Environmental sample testing showed that the LBM environment is highly contaminated with AIVs (Table ). Influenza A detection rates were highest in carcass wash water samples, which serve as ‘pooled’ samples for multiple slaughtered birds. During the slaughtering process at the markets, birds are defeathered and eviscerated before carcasses are washed in a large bucket of water. We observed that 20–30 carcasses were often washed in the same container of water before it was refreshed. These samples were positive for influenza A RNA in 75% of samples from all four markets (ranging from 63 to 86%). Poultry drinking water was also a useful surveillance sample, with 50% of samples testing positive for influenza A RNA; A/H5N1 isolation rates were higher when compared to carcass wash water. Detection rates and isolation rates were also high with soil/mud and discarded feather samples, but these specimens required more complex processing before testing. High contamination of the LBM environment, including relatively high rates of virus isolation, is evidence that the risk of human exposure is very high. Carcass wash water and poultry drinking water samples proved a useful adjunct to poultry swabs for monitoring purposes in the LBMs.
Based solely on the HA typing, analysis of LBM samples collected in 2013 revealed the presence of at least nine other subtypes of AIVs co-circulating with A/H5N1 (Figure 5). This situation increases the likelihood of reassortment events occurring, which may result in the emergence of new influenza subtypes. The recent emergence of reassortant HPAI subtypes of H5, such as A/H5N2, A/H5N5, A/H5N6 and A/H5N8,Citation39, Citation40, Citation41, Citation42, Citation43 is evidence of the risk of new viruses arising through reassortment events with A/H5N1. The sudden emergence of multiple H5 subtypes has not been adequately explained but could be related to the diversity of AIVs currently circulating in domestic poultry populations. The emergence of A/H7N9 virus in China in early 2013Citation44 has also prompted increased concerns about the possibility of a pandemic virus emerging in the Asia-Pacific region. Although most of the non-H5 AIVs detected in this study likely pose little or no risk to humans, the high prevalence of multiple avian influenza strains is an indictment on the poor biosecurity associated with poultry rearing and selling in the region. Previous studies and surveillance have also established that there is intense circulation of poultry between regions and across borders, which facilitates the regional spread of AIVs.Citation9, Citation23
In our study, the detection of A/H5N1 was achieved using qRT-PCR, whereas the detection of other AIVs was done using universal multiplex RT-PCR assays with comparatively reduced sensitivity. In addition, multiplex testing was only conducted where there was disparity between M-gene and H5 qRT-PCR assays. Furthermore, due to the difficulties with classifying influenza viruses using incomplete gene sequences we have only reported LPAI detection where we have been able to generate full HA or NA sequences. It is therefore likely that the true prevalence of non-H5 AIVs was far greater than what we have reported in these market samples. The potential for reassortment between AIVs, including A/H5N1, could result in the emergence of viral strains with considerable impacts on poultry and/or human health. This study, coupled with other recent studies in LBMs from other Asian countries,Citation15, Citation16, Citation17 confirms that the environment of LBMs provides a pool of genes for potential emergence of new pandemic viruses. The silent circulation of a multitude of LPAI viruses, which remain undetected and unmonitored in most Asian countries, heightens the threat when they co-circulate with pandemic candidates such as A/H5N1 and A/H7N9.
As no poultry workers reported any symptoms in relation to acute avian influenza infection, seroconversions in this study would most likely be related to sub-clinical or very mild cases. It has been reported that asymptomatic and mild avian influenza infections lead to seroconversions with low antibody titres that quickly decrease below the threshold recommended by WHO for a confirmed A/H5N1 case (HIA≥160; MN≥80).Citation45 For this reason, we considered as positive all individuals with a HIA titre of ≥80 and a MN titre of ≥40, which is consistentCitation46 or more stringentCitation47, Citation48, Citation49, Citation50, Citation51, Citation52 than the cut-off levels suggested by other authors. However, it is difficult to directly compare the results between studies as there is no consensus on the antibody titre that results from a mild or asymptomatic infection, and consistent cut-off levels have not been used in past studies. According to our classification, 4.5% of workers who participated in the study had antibodies against A/H5N1. This prevalence is much higher than that reported among LBM workers in Egypt,Citation53 ThailandCitation47 or China,Citation54 and much higher than in villagers in ThailandCitation51 or Cambodia.Citation48 The incidence was 3.7 per 1000 person-months in our study, a figure twice that described in Bangladesh in 2009–2010.Citation46 There were no reported clinical signs in our cohort, confirming that a large proportion of human A/H5N1 infections may go undetected. Poultry workers in Cambodia and other Asian countries, where there is endemic circulation of A/H5N1, are constantly exposed to high levels of virus. Although this may lead to mild or sub-clinical infections with seroconversions, it seems that transmission to humans resulting in acute infections is still rare.
Influenza A/H9 viruses, predicted to be A/H9N2 based on the phylogenetic analysis of the HA and NA sequences repeatedly detected in the same samples, were detected in 2.6% of samples analysed in this study. Although A/H9N2 viruses have low pathogenicity for poultry, novel HPAI and LPAI viruses affecting humans, such as A/H5N1, A/H7N9 and A/H10N8, contain internal genes originating from influenza A/H9N2.Citation55, Citation56, Citation57 This may have facilitated the ability of these viruses to cause infection and deaths in humans. Human infections with A/H9N2 have been reported in the literature, and seroprevalence studies have suggested that asymptomatic or mild infections commonly occur in high-risk locations such as LBMs and slaughterhouses.Citation50, Citation53, Citation58, Citation59 The detection of A/H9N2 antibodies in 1.8% of LBM workers participating in our study and an incidence of 0.9 infections per 1000 person-months confirm other data on poultry-to-human transmission of A/H9N2,Citation53, Citation59 including among rural villagers in Cambodia.Citation48 In addition, A/H9N2 infections have also been detected in other mammals such as guinea pigs, dogs, horses and pigs.Citation59, Citation60 The ability of A/H9N2 to cross the species barrier and the relatively high frequency in which it has been implicated in reassortment events suggest that this virus significantly contributes to the emergence of viruses that pose an important public health risk and should therefore be very closely monitored.
In this study we document intense co-circulation of influenza A/H5N1 and LPAI viruses in Cambodian LBMs during 2013. In addition, serological surveys provided evidence of sub-clinical A/H5N1 and A/H9N2 infections. Interventions such as regular cleaning/disinfection, bans on overnight poultry storage, targeted closure during periods of peak circulation and segregation of poultry slaughtering areas should be considered in LBMs to reduce the threat of the emergence of AIVs with public health or animal health impacts. Further monitoring of the circulation of influenza A/H5N1 in Cambodian LBMs and research into the mechanisms associated with human cases is warranted.
Supplementary Table S1
Download PDF (3.6 MB)Supplementary Table S2
Download PDF (4 MB)Supplementary Table S3
Download PDF (3.4 MB)Supplementary Table S4
Download PDF (2.2 MB)Acknowledgments
The authors would like to thank poultry workers at the live-bird markets for their cooperation with the study team. We thank the technical staff at the Virology Unit and investigation team at the Epidemiology and Public Health Unit, Institut Pasteur in Cambodia. The authors would also like to thank the investigation team from the National Veterinary Research Institute, Ministry of Agriculture, Forestry and Fisheries. This work was funded by the Food and Agriculture Organization in Cambodia (Grant NO OSRO/INT/902/USA and OSRO/RAS/901/EC); the office of the Assistant Secretary for Preparedness and Response, US Department of Health and Human Services (Grant NO IDSEP 110011-01-00); and the World Health Organization country office in Cambodia (Grant NO KHM-13-ESR-000137).
Supplementary Information for this article can be found on the Emerging Microbes & Infections website (http://www.nature.com/emi)
- VandegriftKJ,SokolowSH,DaszakPet al.Ecology of avian influenza viruses in a changing world.Ann N Y Acad Sci2010; 1195:113–128.
- CauseyD,EdwardsSV.Ecology of avian influenza virus in birds.J Infect Dis2008; 197:S29–S33.
- WebsterRG,IsachenkoVA,CarterM.A new avian influenza virus from feral birds in the USSR: recombination in nature?Bull World Health Organ1974; 51:325–332.
- SmithGJ,VijaykrishnaD,BahlJet al.Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic.Nature2009; 459:1122–1125.
- KawaokaY,KraussS,WebsterRG.Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics.J Virol1989; 63:4603–4608.
- DucatezMF,HauseB,Stigger-RosserEet al.Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States.Emerg Infect Dis2011; 17:1624–1629.
- TaubenbergerJK.The virulence of the 1918 pandemic influenza virus: unraveling the enigma.Arch Virol2005; 19:101–115.
- YuenKY,ChanPK,PeirisMet al.Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus.Lancet1998; 351:467–471.
- BuchyP,FourmentM,MardySet al.Molecular epidemiology of clade 1 influenza A viruses (H5N1), southern Indochina peninsula, 2004-2007.Emerg Infect Dis2009; 15:1641–1644.
- RithS,DavisCT,DuongVet al.Identification of molecular markers associated with alteration of receptor-binding specificity in a novel genotype of highly pathogenic avian influenza A (H5N1) viruses detected in Cambodia in 2013.J Virol2014; 88:13897–13909.
- The Program for Monitoring Emerging Diseases. Avian influenza (195): Cambodia (BA, SI) H5N1, poultry, OIE. USA: ProMED,2015.Available at:http://www.promedmail.org(archive no. 20151116.3796387).
- WebsterRG.Wet markets—a continuing source of severe acute respiratory syndrome and influenza?Lancet2004; 363:234–236.
- GuanY,SmithGJ.The emergence and diversification of panzootic H5N1 influenza viruses.Virus Res2013; 178:35–43.
- PeirisM,YenHL.Animal and human influenzas.Rev Sci Tech2014; 33:539–553.
- KimKI,ChoiJG,KangHMet al.Geographical distribution of low pathogenic avian influenza viruses of domestic poultry in Vietnam and their genetic relevance with Asian isolates.Poult Sci2013; 92:2012–2023.
- LiJ,YuX,PuXet al.The diversity of avian influenza virus subtypes in live poultry markets before and during the second wave of A(H7N9) infections in Hangzhou, China.Emerg Microbes Infect2015; 4:e14.
- WuH,PengX,PengXet al.Genetic and molecular characterization of H9N2 and H5 avian influenza viruses from live poultry markets in Zhejiang Province, eastern China.Sci Rep2015; 5:17508.
- Van KerkhoveMD,VongS,GuitianJet al.Poultry movement networks in Cambodia: implications for surveillance and control of highly pathogenic avian influenza (HPAI/H5N1).Vaccine2009; 27:6345–6352.
- DeboosereN,HormSV,PinonAet al.Development and validation of a concentration method for the detection of influenza A viruses from large volumes of surface water.Appl Environ Microbiol2011; 77:3802–3808.
- HormSV,DeboosereN,GutiérrezRAet al.Direct detection of highly pathogenic avian influenza A/H5N1 virus from mud specimens.J Virol Methods2011; 176:69–73.
- HormSV,GutiérrezRA,SornSet al.Environment: a potential source of animal and human infection with influenza A (H5N1) virus.Influenza Other Respir Viruses2012; 6:442–448.
- GerloffNA,JonesJ,SimpsonNet al.A high diversity of Eurasian lineage low pathogenicity avian influenza A viruses circulate among wild birds sampled in Egypt.PLoS One2013; 8:e68522.
- SornS,SokT,LySet al.Dynamic of H5N1 virus in Cambodia and emergence of a novel endemic sub-clade.Infect Genet Evol2013; 15:87–94.
- TamuraK,PetersonD,PetersonNet al.MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Mol Biol Evol2011; 28:2731–2739.
- HormSV,SornS,AllalLet al.Influenza A (H5N1) virus surveillance at live poultry markets, Cambodia, 2011.Emerg Infect Dis2013; 19:305–308.
- MountsAW,KwongH,IzurietaHSet al.Case-control study of risk factors for avian influenza A (H5N1) disease, Hong Kong, 1997.J Infect Dis1999; 180:505–508.
- FourniéG,GuitianJ,DesvauxSet al.Identifying live bird markets with the potential to act as reservoirs of avian influenza A (H5N1) virus: a survey in northern Viet Nam and Cambodia.PLoS One2012; 7:e37986.
- NguyenDC,UyekiTM,JadhaoSet al.Isolation and characterization of avian influenza viruses, including highly pathogenic H5N1, from poultry in live bird markets in Hanoi, Vietnam, in 2001.J Virol2005; 79:4201–4212.
- AmonsinA,ChoatrakolC,LapkuntodJet al.Influenza virus (H5N1) in live bird markets and food markets, Thailand.Emerg Infect Dis2008; 14:1739–1742.
- AbdelwhabEM,SelimAA,ArafaAet al.Circulation of avian influenza H5N1 in live bird markets in Egypt.Avian Dis2010; 54:911–914.
- IndrianiR,SamaanG,GultomAet al.Environmental sampling for avian influenza virus A (H5N1) in live-bird markets, Indonesia.Emerg Infect Dis2010; 16:1889–1895.
- MunyuaPM,GithinjiJW,WaibociLWet al.Detection of influenza A virus in live bird markets in Kenya, 2009-2011.Influenza Other Respir Viruses2013; 7:113–119.
- PhanMQ,HenryW,BuiCBet al.Detection of HPAI H5N1 viruses in ducks sampled from live bird markets in Vietnam.Epidemiol Infect2013; 141:601–611.
- ElMasryI,ElshiekhH,AbdlenabiAet al.Avian influenza H5N1 surveillance and its dynamics in poultry in live bird markets, Egypt.Transbound Emerg Dis2015e-pub ahead of print 25 November 2015doi:https://doi.org/10.1111/tbed.12440.
- KungNY,GuanY,PerkinsNRet al.The impact of a monthly rest day on avian influenza virus isolation rates in retail live poultry markets in Hong Kong.Avian Dis2003; 47:1037–1041.
- LauEH,LeungYH,ZhangLJet al.Effect of interventions on influenza A (H9N2) isolation in Hong Kong’s live poultry markets, 1999-2005.Emerg Infect Dis2007; 13:1340–1347.
- LeungYH,LauEH,ZhangLJet al.Avian influenza and ban on overnight poultry storage in live poultry markets, Hong Kong.Emerg Infect Dis2012; 18:1339–1341.
- KungNY,MorrisRS,PerkinsNRet al.Risk for infection with highly pathogenic influenza A virus (H5N1) in chickens, Hong Kong, 2002.Emerg Infect Dis2007; 13:412–418.
- LiuCG,LiuM,LiuFet al.Emerging multiple reassortant H5N5 avian influenza viruses in ducks, China, 2008.Vet Microbiol2013; 167:296–306.
- LeeCC,ZhuH,HuangPYet al.Emergence and evolution of avian H5N2 influenza viruses in chickens in Taiwan.J Virol2014; 88:5677–5686.
- LeeYJ,KangHM,LeeEKet al.Novel reassortant influenza A(H5N8) viruses, South KoreaEmerg Infect Dis2014; 20:1087–1089.
- IpHS,TorchettiMK,CrespoRet al.Novel eurasian highly pathogenic avian influenza a h5 viruses in wild birds, Washington, USA, 2014.Emerg Infect Dis2015; 21:886–890.
- WongFY,PhommachanhP,KalpravidhWet al.Reassortant highly pathogenic influenza A(H5N6) virus in Laos.Emerg Infect Dis2015; 21:511–516.
- GaoR,CaoB,HuYet al.Human infection with a novel avian-origin influenza A (H7N9) virus.N Engl J Med2013; 368:1888–1897.
- BuchyP,VongS,ChuSet al.Kinetics of neutralizing antibodies in patients naturally infected by H5N1 virus.PLoS One2010; 5:e10864.
- NasreenS,KhanSU,LubySPet al.Highly pathogenic Avian Influenza A(H5N1) virus infection among workers at live bird markets, Bangladesh, 2009-2010.Emerg Infect Dis2015; 21:629–637.
- KhuntiratBP,YoonIK,BlairPJet al.Evidence for subclinical avian influenza virus infections among rural Thai villagers.Clin Infect Dis2011; 53:e107–e116.
- BlairPJ,PutnamSD,KruegerWSet al.Evidence for avian H9N2 influenza virus infections among rural villagers in Cambodia.J Infect Public Health2013; 6:69–79.
- ComanA,MafteiDN,KruegerWSet al.Serological evidence for avian H9N2 influenza virus infections among Romanian agriculture workers.J Infect Public Health2013; 6:438–447.
- HuangR,WangAR,LiuZHet al.Seroprevalence of avian influenza H9N2 among poultry workers in Shandong Province, China.Eur J Clin Microbiol Infect Dis2013; 32:1347–1351.
- KruegerWS,KhuntiratB,YoonIKet al.Prospective study of avian influenza virus infections among rural Thai villagers.PLoS One2013; 8:e72196.
- DungTC,DinhPN,NamVSet al.Seroprevalence survey of avian influenza A(H5N1) among live poultry market workers in northern Viet Nam, 2011.Western Pac Surveill Response J2014; 5:21–26.
- GomaaMR,KayedAS,ElabdMAet al.Avian influenza A(H5N1) and A(H9N2) seroprevalence and risk factors for infection among Egyptians: a prospective, controlled seroepidemiological study.J Infect Dis2015; 211:1399–1407.
- HuoX,ZuR,QiXet al.Seroprevalence of avian influenza A (H5N1) virus among poultry workers in Jiangsu Province, China: an observational study.BMC Infect Dis2012; 12:93.
- GuanY,ShortridgeKF,KraussSet al.Molecular characterization of H9N2 influenza viruses: were they the donors of the ‘internal’ genes of H5N1 viruses in Hong Kong?Proc Natl Acad Sci USA1999; 96:9363–9367.
- ChenH,YuanH,GaoRet al.Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study.Lancet2014; 383:714–721.
- PuJ,WangS,YinYet al.Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus.Proc Natl Acad Sci USA2015; 112:548–553.
- PawarSD,TandaleBV,RautCGet al.Avian influenza H9N2 seroprevalence among poultry workers in Pune, India, 2010.PLoS One2012; 7:e36374.
- KhanSU,AndersonBD,HeilGLet al.A systematic review and meta-analysis of the seroprevalence of influenza A (H9N2) infection among humans.J Infect Dis2015; 212:562–569.
- DalbyAR,IqbalM.A global phylogenetic analysis in order to determine the host species and geography dependent features present in the evolution of avian H9N2 influenza hemagglutinin.Peer J2014; 2:e655.