Worldwide dissemination of the mobile colistin-resistance gene mcr-1 is of great concern to public health. Recently, a novel colistin-resistance gene, mcr-3, was reported to share 45.0% nucleotide sequence identity with mcr-1Citation1. To date, mcr-3-positive isolates have been described in several regions (China, Denmark, and Spain) and species (Escherichia coli, Salmonella Typhimurium, and Aeromonas veronii)Citation2–Citation4. Although the plasmid-mediated mcr-3 gene has been identified in most Enterobacteriaceae, with some horizontal mobile elements, such as TnAs2 and ISKpn40Citation1, Citation4, Citation5, no evidence exists of mcr-3 gene transmission mediated by these transposons or insertion sequences. Here, we describe an MCR-3-producing E. coli ST3634 isolate and, for the first time, identify the intermediate circle of the mcr-3-carrying fragment formed by a truncated insertion sequence, IS26, and an intact IS15DI, which was similar to the transfer of mcr-1 via Tn6330Citation6, suggesting this mcr-3-containing segment could be looped out via IS-mediated homologous recombination.
E. coli HN8 was obtained from a fecal sample of an apparently healthy pig at a conventional farm in Henan Province, China, during a routine surveillance study in 2017. PCR and sequencing analyses revealed that mcr-3 was present in E. coli HN8. The mcr-3 in HN8 shared 100% nucleotide sequence identity to the original mcr-3 gene reported by Yin et al.Citation1. Susceptibility was tested by the broth microdilution method following the CLSI guidelines, and the minimal inhibitory concentrations were interpreted from the CLSI breakpointsCitation7. E. coli HN8 was resistant to colistin (4 mg/L), gentamicin (64 mg/L), tetracycline (64 mg/L), ampicillin (64 mg/L), florfenicol ( > 128 mg/L), and trimethoprim/sulfamethoxazole (32/608 mg/L), but susceptible to ceftriaxone (0.03 mg/L), imipenem (0.5 mg/L), aztreonam (0.03 mg/L), ciprofloxacin (0.5 mg/L), and amoxicillin–clavulanic acid (16/8 mg/L).
To further analyze this mcr-3-positive HN8 isolate, whole-cell DNA was extracted using a Wizard genomic DNA purification kit (Promega, Beijing, China) and used for whole-genome sequencing (WGS) on an Illumina Hiseq 2500 platform (Berry Genomics Company, Beijing, China). From the WGS data, sequences types were extracted and assigned to ST3634, and various resistance genes were detected in the WGS data, including aminoglycoside-resistance genes aph(3′)-Ia, aadA5, and aac(3)-IId; rifampicin-resistance gene arr-3; macrolide-resistance gene mph(A); tetracycline-resistance genes tet(A) and tet(M); florfenicol-resistance gene floR, quinolone-resistance genes qnrS1, qnrS2, oqxAB, and aac(6′)Ib-cr; trimethoprim-resistance gene drfA17, β-lactamase-encoding genes blaOXA-1 and blaTEM-1B, tunicamycin-resistance gene tmrB, and bleomycin-resistance gene ble. Four replicons, IncX1, IncR, IncFII, and IncFIB, were detected using PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/).
S1-PFGE and Southern blotting with an mcr-3 probe labeled with digoxigenin revealed that the mcr-3 gene in the HN8 isolate was located on an ~70-kb plasmid, designated pHN8 (data not shown). Although the conjugation failed by filter mating using HN8 as donors and E. coli J53 AzR as the recipient, we obtained the transformant E. coli DH5α/pHN8 by electro-transformation. S1-PFGE and PCR-based plasmid replicon typing analysis indicated the DH5α/pHN8 harbored only the IncR-type plasmid, pHN8. The IncR complex replicons were first described in the K. pneumoniae qnrS1-plasmid, pK245, and carry various resistance genes, including the metallo-β-lactamase genes, blaVIM-1 and blaNDM-1, in clinical Enterobacteriaceae strainsCitation8, Citation9. However, the plasmids belonging to this complex are nontransferable due to the lack of transfer elementsCitation10, which was consistent with our failed conjugation experiment. The transformant DH5α/pHN8 exhibited a 4- to 16-fold increase in the colistin (1 mg/L), gentamicin (32 mg/L), ampicillin (64 mg/L), and ciprofloxacin (0.5 mg/L) MIC values, compared with the recipient DH5α.
The complete pHN8 sequence was obtained by combining the Illumina Hiseq 2500 platform with single-molecule real-time sequencing (SMRT) platforms (Sinobiocore, Beijing, China). pHN8 is a 53,148-bp plasmid, consistent with the size predicted by Southern hybridization within the margin of error, with 66 open reading frames (ORFs) and an average GC content of 49.8%. It contained an 9.14-kb conservative region of the IncR-type plasmid, which shared 98.5% nucleotide sequence identity with that of Enterobacter cloacae plasmid pNDM1_SZ2 (GenBank accession number KU302802) and mainly harbored toxin–antitoxin system gene operon vapB/C, replication gene repB, partition protein-encoding genes parA/B, and SOS mutagenesis gene umuCCitation10. A 32.28-kb MDR region harbored seven of the above-mentioned resistance genes (mcr-3, aac(3)-IId, blaTEM-1B, qnrS1, tet(M), tmrB, and ble), which were flanked by or interspersed with various insertion sequences, with the IS6 family being highly abundant (IS26, n = 4, IS15DI, n = 2, Fig. ).
Fig. 1 a Comparative analysis of pHN8 with closely related plasmids, pWJ1 and pNDM1_SZ2, using the BLAST Ring Image Generator. The concentric rings display similarity between the reference sequence in the inner ring and those in the outer rings. The outermost ring represents resistance gene (red), mobile genetic element (green), and other predicted gene (gray) coding sequences. b Schematic representation of the circular form obtained by sequencing the PCR product and comparing the genetic environment of the pHN8 and pWJ1 mcr-3 genes. The gel picture of the PCR product generated by the Loop-F and Loop-R primers is shown in the top left corner. Lane M: 5000-bp DNA marker; lanes HN8 and DH5α/pHN8: inverse PCR amplicons using HN8 or DH5α/pHN8 as templates. Open reading frames (ORFs) are shown as arrows indicating the transcription direction. △ indicates a truncated gene and the box represents an intact insertion sequence
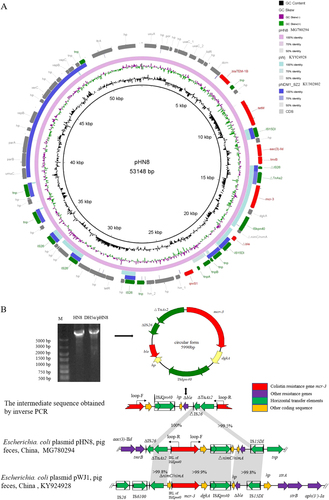
The 6288-bp mcr-3-carrying segment, ranging from △TnAs2 to IS15DI, shared 99.9% nucleotide sequence identity to the corresponding region of the original mcr-3-harboring plasmid, pWJ1 (Fig. ). A truncated (△) IS26 element, the 428-bp 3′-region of IS26 (393–820 bp), was present immediately upstream of △TnAs2. Notably, IS15DI, which differed from IS26 by only two nucleotides (A614G/A615G), was detected downstream of mcr-3. To determine the potential transferability of the pHN8 mcr-3-carrying segment, inverse PCR was performed for both HN8 and its transformant, DH5α/pHN8, using the primers located within the mcr-3 gene (Fig. ). A 5990-bp circular intermediate carrying mcr-3 flanked by △TnAs2, △IS26, and ISKpn40 was identified in both isolates (Fig. ). Based on sequence comparison of plasmid pHN8 and the 5.99-kb circular intermediate, we speculated that it was the two nearly identical insertion sequences, △IS26 and the 3′-region of intact IS15DI, simultaneously contributing to the looping progress by homologous recombination (Fig. ). This event is consistent with previous reports in which △IS26 and the 3′-region of intact IS26 also formed a circular intermediateCitation11, Citation12; however, the mcr-3-carrying circle mobilization evidenced in this study requires further investigation.
In summary, our study describes an MCR-3-producing E. coli ST3634 isolate, and mcr-3 was located on an IncR plasmid that included a mosaic structure containing multiple IS26 sequences. An mcr-3-carrying circular intermediate was mediated by a truncated IS26 and a complete IS15DI, most likely via homologous recombination. Although the pHN8 belongs to an IncR-type non-conjugative plasmid, the mcr-3 gene is transferable either to other plasmids or to chromosomes by IS-mediated transposition, and the IncR plasmid could act as a resistance gene pool in Enterobacteriaceae strains as per previous papersCitation8–Citation10.
Acknowledgements
This study was supported in part by the National Natural Science Foundation of China (31422055 and 81661138002). The plasmid pHN8 nucleotide sequence accession number is MG780294.
Authors' contributions
Y.W. and H.J. designed the study. Z.W. and Y.F. collected the samples and conducted the experiments. Z.W. and Y.W. analyzed and interpreted the data. Z.W., X.-D.D., Y.W., and H.J. drafted the manuscript. All authors reviewed, revised, and approved the final report.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- YinWNovel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coliMBio20178 e00543 175487729
- LitrupEPlasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009-17.Euro Surveill2017223058710.2807/1560-7917.ES.2017.22.31.305875553060
- HernandezMCo-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015Euro Surveill2017223058610.2807/1560-7917.ES.2017.22.31.305865553059
- LingZHChromosome-mediated mcr-3 variants in Aeromonas veroniifrom chicken meatAntimicrob. Agents Chemother.201761e012721710.1128/AAC.01272-17
- LiuLFengYZhangXMcNallyAZongZNew variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5Antimicrob. Agents Chemother.201761e01757175278744
- LiRXieMLvJWai-Chi ChanEChenSComplete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal originJ. Antimicrob. Chemother.201772696699
- CLSI. Performance standards for antimicrobial susceptibility testing. CLSI Supplement 113 M100S 26th edn (Clinical and Laboratory Standards Institute, Wayne, PA, 2016).
- PapagiannitsisCCMiriagouVGiakkoupiPTzouvelekisLSVatopoulosACCharacterization of pKP1780, a novel IncR plasmid from the emerging Klebsiella pneumoniae ST147, encoding the VIM-1 metallo-beta-lactamaseJ. Antimicrob. Chemother.2013682259226210.1093/jac/dkt196
- KocsisEblaNDM-1 carriage on IncR plasmid in Enterobacteriaceae strainsMicrob. Drug Resist.20162212312810.1089/mdr.2015.0083
- CompainFComplete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniaeAntimicrob. Agents Chemother.2014584207421010.1128/AAC.02773-134068571
- HarmerCJMoranRAHallRMMovement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26MBio20145e01801e0181410.1128/mBio.01801-144196232
- HarmerCJHallRMIS26-mediated precise excision of the IS26-aphA1a translocatable unitMBio20156e018661510.1128/mBio.01866-154676283
