Abstract
Microsporidia cause opportunistic infections in highly immunodeficient individuals. Few studies on the epidemiology of these infections have been conducted in France. Between 2014 and 2016, we undertook a study to estimate the prevalence and circulating genotypes of this fungus-related micro-organism among the population of Strasbourg University Hospital. Samples were collected from hospitalized patients and analyzed using microscopy and molecular assays. Strains from positive subjects were sequenced for genotyping. Only 7/661 patients (1.1%) were positive for microsporidia, and the only species identified was Enterocytozoon bieneusi. Two patients presented immunodeficiency linked to AIDS, and five transplant recipients presented immunodeficiency linked to immunosuppressive therapies. Only five patients received specific antimicrosporidial treatment, but clinical outcomes were good in all cases. We identified four genotypes: A and D in patients with AIDS, and C and S9 in transplant recipients. To date, genotype S9 has been described only once. This genotype is similar to those found in farm animals in China. Because some of these animals have been introduced to Central Europe, we postulate that this genotype might be of Asian origin. Thus, genotyping microsporidial strains may be of epidemiological and clinical interest to identify potential outbreak sources depending on the infecting strains.
Introduction
Microsporidia are a group of unicellular fungi living as obligate intracellular parasitesCitation1. The first microsporidial species to be described, Nosema bombycis, was identified by Carl Wilhelm von Nägeli in 1857Citation2 as the causative agent of a silkworm disease. Since then, approximately 1000–1500 microsporidial species have been describedCitation1. Over the past 150 years, microsporidia classification has been widely discussed, based on morphological, biochemical, and genetic characteristics. Although NägeliCitation2 initially placed Nosema bombycis in the fungal group, Schizomycetes, microsporidia were considered protozoans for more than a centuryCitation1. Recently, phylogenetic analyses of five gene sequences (mitochondrial HSP70, TATA-box protein, RNA polymerase II, and α-tubulin and β-tubulin) have supported classifying microsporidia within the fungi or as a sister-group with a common ancestorCitation1.
Until the 1970s, microsporidial species were rarely recognized as causes of human pathologyCitation1. With the HIV epidemic emerging in the first half of the 1980s, microsporidia, which cause opportunistic infections in highly immunodeficient individuals, became more frequent and visible, concomitant with physicians’ increasing knowledge of this conditionCitation1. Thus, diverse symptoms were associated with microsporidial infections, most commonly digestive disorders, but many other unusual symptoms as wellCitation1. Four species primarily cause human infections: Enterocytozoon bieneusi, Encephalitozoon intestinalis, Encephalitozoon hellem, and Encephalitozoon cuniculi.
Few studies on the epidemiology of these infections have been conducted in France, and most of these studies were on microsporidial infections in HIV-infected patients. However, current patients undergoing immunosuppressive therapies, such as transplant recipients, account for many of the immunocompromised patients, and microsporidial infections have been described under these conditionsCitation3. In this retrospective study, we evaluated the frequency of these infections in Eastern France by analyzing cases diagnosed between January 2014 and December 2016 at the Strasbourg University Hospital. We were interested in epidemiological, clinical, and microbiological features, including genotyping, to determine the current burden and characteristics of these infections in Eastern France.
Results
Clinical features
Between January 2014 and December 2016, 661 samples were sent to the laboratory of the University Hospital of Strasbourg for microsporidia testing. Of these, 613 (92.7%) came from immunocompromised patients, including most organ transplant recipients (61.1%) (Table ). Samples were taken from 281 females (42.5%) and 380 males, with an average age of 52.27 years (range 16 days–92 years), and 57 samples (8.6%) were from patients under 18-years old. Only seven patients (1.1%) were positive for microsporidia, and the only species detected was E. bieneusi. The positive individuals’ ages ranged from 7 to 63 years, and there were 5 males and 2 females (Table ).
Underlying conditions of patients tested for intestinal microsporidiosis
Patients positive for E. bieneusi
All patients were immunodeficient; two patients were linked to HIV infection with AIDS (C3 stage), and the other five were linked to immunosuppressive therapies. The two patients with HIV had very low CD4 counts (38 and 61/mm3). For patients undergoing immunosuppressive therapies, these treatments were administered to prevent organ rejection after transplantation (four kidneys, one heart), and all five patients were receiving at least a 2-drug regimen, including mycophenolate, tacrolimus, everolimus, ciclosporin, and prednisone (Table ).
All seven patients presented digestive symptoms with acute diarrhea (three patients), chronic diarrhea (three patients), or “yellow” stool (one patient). Six patients presented with biological inflammatory syndrome with moderately elevated C-reactive protein (CRP) ranging from 24.5 mg/l to 109 mg/l. All positive patients presented comorbidities (tuberculosis, hepatitis B and C, pneumonia, pericarditis, pneumocystosis, macrophage activation syndrome, cryptococcosis, and transplant rejection).
Five patients received specific antimicrosporidial treatment with four regimens (Table ). We could not access treatment information for one patient because of incomplete medical records, and two patients received no specific treatment for the E. bieneusi infection; however, these three subjects were treated for other infections: one with azithromycin, one with azithromycin and trimethoprim-sulfamethoxazole, and the last with spiramycin, cefotaxime, vancomycin, and valganciclovir.
The clinical outcomes were good with digestive symptoms being resolved in all cases. Three patients received a second parasitological control examination 2–6 weeks after diagnosis. Two remained positive at this examination.
Microbiological features
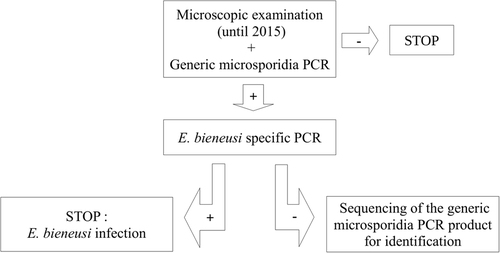
Microsporidia was diagnosed using two polymerase chain reactions (PCRs), the first to detect microsporidial species from the Enterocytozoon and Encephalitozoon genera and the second specific for E. bieneusi onlyCitation4 (Fig. ). All subjects positive for microsporidia were infected with E. bieneusi. Of the 661 samples examined, 46 (6.9%) were positive for other parasites. Only one (subject 5) was coinfected with E. bieneusi, Blastocystis spp., and Chilomastix mesnili. Eight patients were coinfected with 2–3 parasites but not E. bieneusi. All other patients positive for parasites were mono-infected (detailed in Table ).
All parasite results from 661 patient stool samples
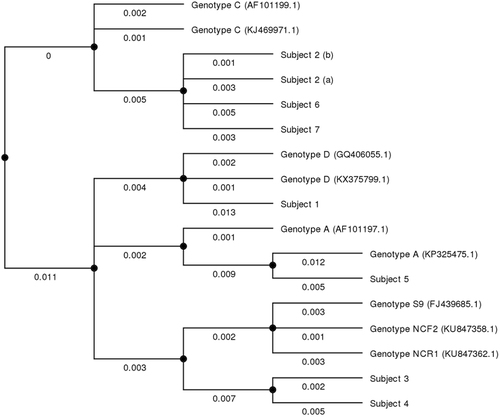
To genotype the isolated strains, another PCR was performed using primers targeting internal transcribed spacers (ITS) between small-subunit rRNA and large-subunit rRNA genesCitation5. This procedure allowed comparison with published sequences in the GenBank database. All sequences were genetically similar, but four genotypes were identified (Fig. ). One HIV-positive patient was infected with E. bieneusi genotype D, the other with genotype A. Among the five patients receiving immunosuppressive therapies, three were infected with E. bieneusi genotype C (all were kidney-transplant recipients) and two with genotype S9 (one kidney transplant and one heart transplant recipient). Subject 2 tested positive twice for genotype C. Subject 5 tested positive twice for E. bieneusi, with genotype A for the first sample; however, the volume of the second sample was insufficient and could not be genotyped.
Numbers under the lines show relative distances between the strains. Study patients are marked 1–7. Reference sequences were those used by Santín et al. for E. bieneusi nomenclature consensusCitation26 and are marked with their GenBank accession number
Discussion
We report seven cases of intestinal microsporidiosis due to E. bieneusi in immunodeficient patients. Most patients were transplant recipients with marked immunodeficiency mediated by anti-organ-rejection therapies. With the development of HIV protease inhibitors in 1995Citation6 and the use of combined antiretroviral therapy (cART)Citation7, subjects with HIV have become less susceptible to microsporidial diseaseCitation8–Citation12. More recently, powerful immunosuppressive therapeutics were used to prevent organ rejection after transplantationCitation13, putting transplant recipients at risk of infectionCitation14. In addition, HIV-positive patients with very low CD4+ cell counts remain at risk. All patients who were screened for microsporidia presented symptoms attributable to E. bieneusi infection, which is known to cause a relatively localized disease, with a tropism for the gastrointestinal tractCitation1. Diagnoses were performed using microscopy (until 2015) and PCR; however, this search was positive only 10 times for 7 patients over 3 years, with 661 tests performed. Thus, microsporidial disease seems to be an uncommon diagnosis at our hospital.
Only four of the seven diagnosed patients were specifically treated for E. bieneusi infection, with four drug regimens using nitazoxanide and/or albendazole. Two patients remained untreated and we found no information on the last patient. Outcomes were good in all cases. Currently, France has no official treatment recommendation for intestinal microsporidiosis or E. bieneusi infection; the most commonly used drugs are nitazoxanideCitation15, Citation16 and albendazoleCitation17, Citation18. However, several successful treatments have been reported using fumagillin for intestinal microsporidiosis due to E. bieneusi in transplant recipientsCitation19–Citation23. Furthermore, in most previously reported cases, immunosuppressive therapies were tapered or discontinued at the same time treatment was introducedCitation20. Similarly, in our study, cART was initiated in HIV-infected patients, and immunosuppressive therapies were tapered in transplant recipients. Since introducing cART, many positive outcomes have been reported for intestinal microsporidiosis after immune restoration, with no other specific treatmentCitation8–Citation12. However, a recent publication showed that intestinal microsporidiosis due to E. bieneusi in a stem cell transplant recipient was successfully treated with fumagillin and avoided modifications of immunosuppressionCitation23. In any case, patient immunity seems to be a key element in curing this infection. Finally, given the low prevalence of E. bieneusi infections in patients with diarrhea in our study, directly implicating this microorganism in intestinal symptom development in these patients remains uncertain. All samples analyzed were from patients experiencing diarrhea, but only 7 (1.1%) were positive for E. bieneusi. These patients may have only been carriers of this microorganism. These elements raise the question of the value of diagnosing intestinal microsporidiosis and using specific treatments.
We identified E. bieneusi strains that were genetically similar to four described genotypes: A, C, D, and S9. The most predominant genotype was C, found in three patients, all of whom were kidney transplant recipients. Genotype C is the most common genotype in kidney transplant patients, with a high specificity for this immune backgroundCitation5, Citation24, as well as for liver transplant recipientsCitation25. We described one patient infected with genotype A and one with genotype D, both being HIV-infected with AIDS. These genotypes are frequently observed in HIV-infected patientsCitation5; however, we did not isolate the most commonly described genotype in this HIV setting, genotype BCitation5, Citation26, Citation27. The fourth genotype was S9, carried by two patients undergoing immunosuppressive therapies, one being a heart transplant recipient, the other a kidney transplant recipient. To our knowledge, this genotype has only been described once, in a patient with ulcerative colitis in the NetherlandsCitation5. This genotype is very similar to two other recently described genotypes, NCR1 and NCF2, found in raccoon dogs (Nyctereutes procyonoides) farmed in Northern ChinaCitation28. Other highly similar strains to these three genotypes have mostly been found in animals in ChinaCitation29–Citation31 and recently in wild animals, especially wild boars (Sus scrofa) and raccoons (Procyon lotor) introduced in Central EuropeCitation32, Citation33. With Strasbourg being on the German border, these strains are likely to circulate in our region, possibly after having been introduced from Asia. It would be interesting to genotype more strains isolated from patients from the Rhine valley and Central Europe to assess whether these strains occur frequently in those areas.
Furthermore, outbreaks related to environmental contamination can occur in immunodeficient and immunocompetent individualsCitation34. Although infections are believed to occur via ingestionCitation1, Citation35, E. bieneusi can infect other mammals, birds, insects, and arthropods, making the contamination source difficult to ascertain, with a potential for zoonotic and arthropod-borne transmissionCitation18. Thus, this makes it difficult to recommend preventive behaviors to patients potentially at risk of microsporidial infection, and strain genotyping should be routinely performed to identify environmental sources that may cause outbreaks.
In conclusion, given its epidemiological characteristics linked to immunocompromised hosts, intestinal microsporidiosis due to E. bieneusi is an uncommon but potentially emerging disease with increased use of immunosuppressive treatments. Our study shows that, in our region, transplant recipients are the main population at risk of infection. The absence of treatment recommendations leaves physicians with the choice of a drug regimen that does not appear to affect the usually favorable course of infection, and patient immunity might be a key element in triggering and curing this infection. In this context, genotyping strains isolated from patients may help identify environmental sources that could potentially initiate outbreaks. Finally, we identified strains similar to uncommon genotypes that may have been introduced through animals imported from Asia. This would show the importance of human activities in spreading human and animal pathogens.
Materials and methods
Sample collection
All samples were collected from patients at the University Hospital of Strasbourg per standard procedures over a 3-year period (January 2014 to December 2016). Subjects were included when their clinician specifically requested testing for microsporidia.
Microscopic examination
A first microscopic examination was performed without staining shortly after stool collection. Samples were then examined after merthiolate-iodine-formaldehyde staining and concentration (Faust-Ingalls method). From January to December 2014, all samples were also stained with Weber’s modified trichrome for microsporidial spore staining before microscopic examination by a trained parasitologist. This step was discontinued in 2015 due to a change in legislation prohibiting use of the toxic reagents necessary for Weber’s staining and because no superiority in sensitivity or specificity has been found compared with PCRCitation36.
Molecular assays
An aliquot of each stool sample was stored at −20 °C until molecular diagnosis. DNA was extracted using the DNA Stool minikit (Qiagen, the Netherlands) per the manufacturer’s recommendations, without prior mechanical disruption. All samples were submitted to a first PCR using a commercial “microsporidia generic” real-time PCR kit (Bio-Evolution, France) to detect all species belonging to the Enterocytozoon and Encephalitozoon genera and amplifying a sequence of the 18S rRNA gene (Se = 88%, Sp = 83%) per the manufacturer’s recommendations. Positive samples were submitted to a second PCR specific for E. bieneusi. We used the primers Eb.gc (5′-TCAGTTTTGGGTGTGGTATCGG-3′) and Eb.gt (5′-GCTACCCATACACACATCATTC-3′), which amplify a 210-bp DNA fragment in ITS sequence of E. bieneusi, as previously describedCitation4. All PCR reactions were performed using GoTaq (Promega, USA) polymerase. To genotype the different strains found in the samples, we performed a third PCR on the ITS sequence (GenBank accession no. AF101200). The forward primer, Eb-80F (5′-GTTGGAGAACCAGCTGAAGGT-3′), and reverse primer, Eb-375R (5′-ATACACCTCTTGATGGCACCCT-3′), amplified a 296-bp fragment, as previously describedCitation5. The latter PCR product was sent for sequencing (GATC Biotech AG, Germany).
Sequences obtained were compared with the GenBank database (www.ncbi.nlm.nih.gov/genbank/) and aligned to draw a phylogenic tree comparing the sequences. This tree was designed with the MrBayes 3.2 phylogenetic programCitation37 performed through the open-source bioinformatics software, UGENE (Unipro, Russia)Citation38.
Data availability
All data supporting these findings are available without restriction upon request to the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- FranzenCMicrosporidia: a review of 150 years of researchOpen Parasitol. J.20082 1 3410.2174/1874421400802010001
- NageliKWUber die neue Krankheit der Seidenraupe und verwandte OrganismenBot. Z185715760761
- DidierESWeissLMMicrosporidiosis: not just in AIDS patientsCurr. Opin. Infect. Dis.20112449049510.1097/QCO.0b013e32834aa1523416021
- LiguoryODavidFSarfatiCDerouinFMolinaJMDetermination of types of Enterocytozoon bieneusi strains isolated from patients with intestinal microsporidiosisJ. Clin. Microbiol.19983618821885104945
- ten HoveRJCharacterization of genotypes of Enterocytozoon bieneusi in immunosuppressed and immunocompetent patient groupsJ. Eukaryot. Microbiol.20095638839310.1111/j.1550-7408.2009.00393.x
- DannerSAA short-term study of the safety, pharmacokinetics, and efficacy of ritonavir, an inhibitor of HIV-1 protease. European-Australian Collaborative Ritonavir Study GroupN. Engl. J. Med.19953331528153310.1056/NEJM199512073332303
- AutranBPositive effects of combined antiretroviral therapy on CD4+T cell homeostasis and function in advanced HIV diseaseScience199727711211610.1126/science.277.5322.112
- GoguelJRemission of AIDS-associated intestinal microsporidiosis with highly active antiretroviral therapyAIDS19971116581659
- CarrAMarriottDFieldAVasakECooperDATreatment of HIV-1-associated microsporidiosis and cryptosporidiosis with combination antiretroviral therapyLancet199835125626110.1016/S0140-6736(97)07529-6
- ConteasCNModification of the clinical course of intestinal microsporidiosis in acquired immunodeficiency syndrome patients by immune status and anti-human immunodeficiency virus therapyAm. J. Trop. Med. Hyg.19985855555810.4269/ajtmh.1998.58.555
- FoudraineNAImprovement of chronic diarrhoea in patients with advanced HIV-1 infection during potent antiretroviral therapyAIDS199812354110.1097/00002030-199801000-00005
- MiaoYMEradication of cryptosporidia and microsporidia following successful antiretroviral therapyJ. Acquir. Immune Defic. Syndr.20002512412910.1097/00126334-200010010-00006
- Meier-KriescheHUImmunosuppression: evolution in practice and trends, 1994-2004Am. J. Transplant.200661111113110.1111/j.1600-6143.2006.01270.x
- FishmanJARubinRHInfection in organ-transplant recipientsN. Engl. J. Med.19983381741175110.1056/NEJM199806113382407
- DidierESScreening of compounds for antimicrosporidial activity in vitroFolia Parasitol.199845129139
- Bicart-SéeAMassipPLinasMDDatryASuccessful treatment with nitazoxanide of Enterocytozoon bieneusi microsporidiosis in a patient with AIDSAntimicrob. Agents Chemother.20004416716810.1128/AAC.44.1.167-168.200089645
- BlanshardCEllisDSToveyDGDowellSGazzardBGTreatment of intestinal microsporidiosis with albendazole in patients with AIDSAIDS1992631131310.1097/00002030-199203000-00009
- DieterichDTLewEAKotlerDPPolesMAOrensteinJMTreatment with albendazole for intestinal disease due to Enterocytozoon bieneusi in patients with AIDSJ. Infect. Dis.199416917818310.1093/infdis/169.1.178
- MolinaJMPotential efficacy of fumagillin in intestinal microsporidiosis due to Enterocytozoon bieneusi in patients with HIV infection: results of a drug screening study. The French Microsporidiosis Study GroupAIDS1997111603161010.1097/00002030-199713000-00009
- ChampionLFumagillin for treatment of intestinal microsporidiosis in renal transplant recipientsAm. J. Transplant.2010101925193010.1111/j.1600-6143.2010.03166.x
- GodronAAccoceberryICouretALlanasBHarambatJIntestinal microsporidiosis due to Enterocytozoon bieneusi in a pediatric kidney transplant recipient successfully treated with fumagillinTransplantation201396e66e6710.1097/TP.0b013e3182a902e7
- DesoubeauxGSuccessful treatment with fumagillin of the first pediatric case of digestive microsporidiosis in a liver-kidney transplantTranspl. Infect. Dis.201315E250E25910.1111/tid.12158
- BukreyevaIEnterocytozoon bieneusi microsporidiosis in stem cell transplant recipients treated with fumagillinEmerg. Infect. Dis.2017231039104110.3201/eid2306.1618255443440
- LanternierFMicrosporidiosis in solid organ transplant recipients: two Enterocytozoon bieneusi cases and reviewTranspl. Infect. Dis.200911838810.1111/j.1399-3062.2008.00347.x
- SingATybusKHeesemannJMathisAMolecular diagnosis of an Enterocytozoon bieneusi human genotype c infection in a moderately immunosuppressed human immunodeficiency virus- seronegative liver-transplant recipient with severe chronic diarrheaJ. Clin. Microbiol.2001392371237210.1128/JCM.39.6.2371-2372.200188153
- SantínMFayerREnterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensusJ. Eukaryot. Microbiol.200956343810.1111/j.1550-7408.2008.00380.x
- LiguoryOSarfatiCDerouinFMolinaJMEvidence of different Enterocytozoon bieneusi genotypes in patients with and without human immunodeficiency virus infectionJ. Clin. Microbiol.2001392672267410.1128/JCM.39.7.2672-2674.200188208
- XuCPrevalence, risk factors and molecular characterization of Enterocytozoon bieneusi in raccoon dogs (Nyctereutes procyonoides) in five provinces of Northern ChinaActa Trop.2016161687210.1016/j.actatropica.2016.05.015
- ZhangZZoonotic Enterocytozoon bieneusi genotypes in Pere David’s deer (Elaphurus davidianus) in Henan, ChinaExp. Parasitol.2015155464810.1016/j.exppara.2015.05.008
- LiJMolecular characterization of Cryptosporidium spp., Giardiaduodenalis, and Enterocytozoonbieneusi in captive wildlife at Zhengzhou Zoo, ChinaJ. Eukaryot. Microbiol.20156283383910.1111/jeu.12269
- ZhangXXPrevalence, risk factors and multilocus genotyping of Enterocytozoonbieneusi in farmed foxes (Vulpeslagopus), Northern ChinaParasit. Vectors201697210.1186/s13071-016-1356-14743323
- LeśniańskaKCryptosporidium spp. and Enterocytozoonbieneusi in introduced raccoons (Procyonlotor)-first evidence from Poland and GermanyParasitol. Res.20161154535454110.1007/s00436-016-5245-55104802
- NěmejcKPrevalence and diversity of Encephalitozoon spp. and Enterocytozoonbieneusi in wild boars (Susscrofa) in Central EuropeParasitol. Res.201411376176710.1007/s00436-013-3707-6
- CotteLPrevalence of intestinal protozoans in French patients infected with HIVJ. Acquir. Immune Defic. Syndr.1993610241029
- MathisAWeberRDeplazesPZoonotic potential of the microsporidiaClin. Microbiol. Rev.20051842344510.1128/CMR.18.3.423-445.20051195965
- SubrungruangIEvaluation of DNA extraction and PCR methods for detection of Enterocytozoonbieneusi in stool specimensJ. Clin. Microbiol.2004423490349410.1128/JCM.42.8.3490-3494.2004497608
- RonquistFMrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model spaceSyst. Biol.20126153954210.1093/sysbio/sys0293329765
- OkonechnikovKGolosovaOFursovMUnipro UGENE: a unified bioinformatics toolkitBioinforma2012281166116710.1093/bioinformatics/bts091