Yersinia enterocolitica, a member of the Enterobacteriaceae family, is a gastrointestinal pathogen. This species is transmitted via the fecal-oral route, usually through the consumption of contaminated food, and less often through direct contact with infected animalsCitation1. Y. enterocolitica infections cause diarrhea, fever, and abdominal pain, mimicking appendicitis, and predominate in childrenCitation2. The species Y. enterocolitica is subdivided into six biotypes: the nonpathogenic biotype 1A, low-pathogenicity biotypes 2–5, which cause self-limiting infections, and the highly pathogenic biotype 1B, which is more commonly associated with systemic disseminationCitation3,Citation4.
While biotype 4 is now predominant in North AmericaCitation1,Citation5, biotype 1B was responsible for most of the Y. enterocolitica infections in the USA until the end of the 1980sCitation5,Citation6. The first outbreak due to Y. enterocolitica 1B was reported in the state of New York, USA, in 1976, with more than 200 children and school employees infected after the consumption of contaminated chocolate milk, resulting in hospitalization of 36 childrenCitation7. Other outbreaks were reported in 1981, 1982, and 1995, due to the consumption of powder milk, tofu, or Pasteurized milkCitation1.
Y. enterocolitica biotype 1B strains have rarely been isolated in Europe. In the past 20 years, only two clinical strains of this biotype have been isolated in France among a total of 9442 Yersinia strains characterized by the French National Reference Laboratory, with the last one, isolated from a clinical cutaneous abscess, dating to 2004 (unpublished data). In Italy, five biotype 1B strains were isolated between 1979 and 1989Citation8, and in Germany, the first isolation was reported in 2002Citation9, but no isolations have been reported in these countries since then. On the other hand, the first Y. enterocolitica 1B strain in Poland was isolated in 2004 from a patient who suffered from liver cirrhosisCitation10, and the number of isolated strains has dramatically increased since thenCitation11,Citation12. Owing to the systemic dissemination potential of Y. enterocolitica 1B strains, their spread may cause a serious public health threat, thus rendering their surveillance crucial.
Herein, we report the case of Y. enterocolitica 1B infection in a French patient, as well as the characterization of the strain and genomic analysis of its genetic relatedness to other biotype 1B strains.
The case
Here, we report the case of a French 42-year-old woman who presented with chronic diarrhea, abdominal pain, and vomiting in August 2017. No history of travel outside metropolitan France in the last 6 months was reported. She was treated with ciprofloxacin for 7 days. Subsequently, the patient developed inflammatory bowel disease, and a colonoscopy revealed terminal ileitis, a hallmark of enteric yersiniosisCitation13. Collected stools were semiliquid and glairy, and their microscopic examination revealed the presence of rare leukocytes but no erythrocytes, suggesting a bacterial infection. A stool sample was seeded on cefsulodin irgasan novobiocin (CIN) agar medium (Biomérieux, Cat. #43421, Craponne, France), and incubated for 48 h at 30 °C. Few Yersinia-like colonies grew, and a Y. enterocolitica strain was identified by mass spectrometry (MALDI-TOF, Bruker Microflex, BDD 6903 MSP) with a score of 2.37. The strain was sent to the French Yersinia National Reference Laboratory (Institut Pasteur, Paris, France), where tests using API20E and 50CH strips, Tween-esterase, pyrazinamidase activity, and O-antigen seroagglutination revealed that the strain (IP39285) belonged to the Y. enterocolitica bioserotype 1B/O:7,8-8-8,19. Since isolation of biotype 1B strains is extremely rare in France, polymerase chain reaction (PCR), targeting yadA (located on the Yersinia virulence plasmid) and fyuA (located in the Yersinia high-pathogenicity island), was performed. Both PCR tests were positive, confirming that IP39285 was a highly pathogenic biotype 1B strain. Multilocus sequence analysis (glnA, gyrB, hsp60, and recA)Citation14 showed that this strain belonged to the same branch as other biotype 1B strains (data not shown). Antimicrobial susceptibility testing, based on the 2017 EUCAST guidelines, revealed resistance of IP39285 to amoxicillin, as usually observed for Y. enterocolitica 1B strains, and its susceptibility to all other antibiotics tested (amoxicillin-clavulanic acid, cephalexin, cefoxitin, ceftriaxone, ciprofloxacin, nalidixic acid, trimethoprim, sulfonamide, tetracycline, and ticarcillin).
We next evaluated the genetic relatedness of IP39285 to three other biotype 1B strains of French, Belgian, and USA origin previously isolated at the Yersinia National Reference Laboratory, and to 16 biotype 1B strains of Belgian, German, USA and unknown origin, whose genomic sequences are publicly available (see Supplementary Table). Whole-genome sequencing of strains IP39285, IP28308, IP35698, and IP38642 was performed using a NextSeq® 500 sequencing system (Illumina, San Diego, CA, USA). The annotated genome of the IP39285 strain was deposited to GenBank, and is available under BioProject PRJNA407758 (accession number NWMR00000000). The genome was annotated with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP), and consists of 4,325,608 bp in 143 contigs, with a G + C content of 47.5%. PGAP found a total of 4061 genes, 3988 coding sequences (CDSs), 73 RNA genes, and 169 pseudogenes. The G + C content and the number of CDSs are similar to those of other publicly available genomes of biotype 1B Y. enterocolitica strains. In silico genomic analysis revealed the presence of classic virulence genes, including ail, inv, ystA, and myfA, located on the chromosome; fyuA and irp2, located in the Yersinia high-pathogenicity island; and yadA and virF, located on the Yersinia virulence plasmid. Prediction of antimicrobial susceptibility genes in the IP39285 genome using ResFinder (http://www.genomicepidemiology.org/) allowed the identification of the blaA gene, responsible for beta-lactam resistance, as in all other biotype 1B strains, and of the vatF gene, responsible for streptogramin B resistance, as in 14 out of the 18 other biotype 1B genomes tested.
We then conducted a comparative genomic analysis to identify single-nucleotide polymorphisms (SNPs) with the wgSNP analysis module of Bionumerics 7.6 (Applied Maths, Sint-Martens-Latem, Belgium) using the genome of strain 8081 as a reference (accession number AM286415) (Fig. ). In total, 25,671 SNP positions were identified in the 19 compared strains, highlighting a high genetic diversity among these isolates (Fig. ). For comparison, in 264 Y. enterocolitica biotype 4 strains with no epidemiological links isolated worldwide in the past 30 years, only 9827 SNPs were identified (data not shown). This diversity was also observed using a pairwise comparison with numerous branches, whose length was more than 1000 SNPs. Despite this diversity, we identified a cluster of six highly homogeneous strains (maximum difference of approximately 100 SNPs), which included IP39285. Within this cluster, a Belgian strain, IP38642, and German strains, SZ506/04, SZ375/34, and SZ5108/01, had fewer than 18 SNPs, indicating that the strains were highly related to each other. The USA strain Y286 had only a 37-SNP difference from the German strain SZ375/04, despite their geographical distance. The closest relative to the French strain IP39285 was the Y286 strain, with a 95-SNP difference. Other European and North American strains were distant from the Y286 strain, with at least 2356 SNP difference.
Numbers on the branches indicate the difference in SNPs between two strains. Logarithmic scaling was used for branch lengths
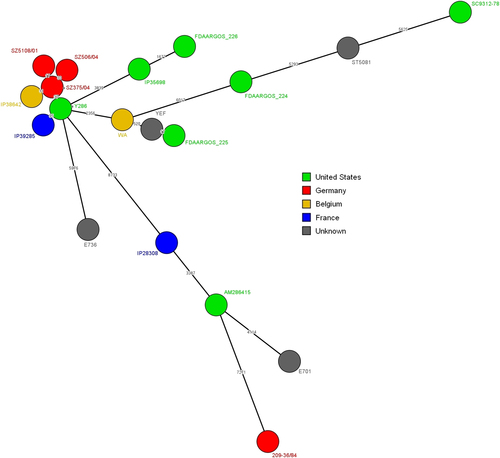
Altogether, analysis of the cluster including strain IP39285 and of the other, more distant branches suggested potential circulation of Y. enterocolitica biotype 1B strains between North America and Europe. We were also able to detect the microevolution of a Y. enterocolitica biotype 1B clone settled in Europe, including Belgian and German strains. Considering the recent increase in Y. enterocolitica biotype 1B infections in some European countries, our results highlight the need for more sustained surveillance of this highly pathogenic strain, whose spread may pose a global public health threat.
Supplementary Table S1
Download MS Word (13.4 KB)Acknowledgements
We would like to thank Dr. Nathalie Oleron (Chalezeule, France) who provided clinical information about the patient, and Dr. Marc Lecuit (Institut Pasteur, Paris, France) for critical reading of the manuscript. This work was funded by Institut Pasteur (Paris, France) and by Santé Publique France (Saint-Maurice, France).
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0123-0).
References
- Carniel, E. et al. in The Prokaryotes (eds Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E.) Y. enterocolitica and Y. pseudotuberculosis, pp. 270–398. (Springer-Verlag, New York, 2006).
- RosnerBMWerberDHohleMStarkKClinical aspects and self-reported symptoms of sequelae of Yersinia enterocolitica infections in a population-based study, Germany 2009-2010BMC Infect. Dis.20131310.1186/1471-2334-13-2363669037
- ReuterSParallel independent evolution of pathogenicity within the genus YersiniaProc. Natl. Acad. Sci. USA2014111 6768 677310.1073/pnas.1317161111
- WautersGKandoloKJanssensMRevised biogrouping scheme of Yersinia enterocoliticaContrib. Microbiol. Immunol.198791421
- BissettMLPowersCAbbottSLJandaJMEpidemiologic investigations of Yersinia enterocolitica and related species: sources, frequency, and serogroup distributionJ. Clin. Microbiol.199028910912267835
- KayBAVirulence and phenotypic characterization of Yersinia enterocolitica isolated from humans in the United StatesJ. Clin. Microbiol198317128138272587
- BlackREEpidemic Yersinia enterocolitica infection due to contaminated chocolate milkN. Engl. J. Med1978298767910.1056/NEJM197801122980204
- MartiniAFantasiaMRaballoAYersinia enterocolitica 08 “American strain” isolated in ItalyMicrobiologica198912345347
- SchubertSBockemuhlJBrendlerUHeesemannJFirst isolation of virulent Yersinia enterocolitica O8, biotype 1B in GermanyEur. J. Clin. Microbiol Infect. Dis.2003226668
- Szych, J. et al. First isolation of virulent Yersinia enterocolitica O8, biotype 1B in Poland. Post Mikrobiol43, (Suppl 1), 159 (2004).
- RastawickiWSzychJGierczynskiRRokoszNA dramatic increase of Yersinia enterocolitica serogroup O:8 infections in PolandEur. J. Clin. Microbiol Infect. Dis.20092853553710.1007/s10096-008-0647-7
- RastawickiWSzychJRokoszNZacharczukKGierczynskiRSeasonality of Yersinia enterocolitica bioserotype 1B/O:8 infections in PolandEpidemiol. Infect.20131412039204210.1017/S0950268812002786
- KatoYHattoriTOh-YaHYoshinoSKatoHAcute terminal ileitis and Yersinia enterocolitica infectionGastroenterol. Jpn.197712364310.1007/BF02774000
- KotetishviliMMultilocus sequence typing for studying genetic relationships among Yersinia speciesJ. Clin. Microbiol.2005432674268410.1128/JCM.43.6.2674-2684.20051151872
