Abstract
This study investigated the characteristics and mechanisms of eravacycline resistance and heteroresistance in clinical Klebsiella pneumoniae isolates. A total of 393 clinical K. pneumoniae isolates were collected and subjected to eravacycline and tigecycline MIC determinations using the agar dilution method. Eravacycline heteroresistance was assessed by a population analysis profile (PAP). The expression levels of efflux pumps and their regulators were determined by quantitative reverse-transcription PCR (qRT-PCR). This study identified 67 eravacycline-nonsusceptible isolates; among the extended-spectrum β-lactamase (ESBL)-positive isolates, eravacycline-nonsusceptible isolates were detected more frequently than tigecycline-nonsusceptible isolates (21.7% vs. 9.4%, p = 0.001). The study sample was observed to include 20 K. pneumoniae isolates with eravacycline heteroresistance. Compared to the reference strain, oqxA or oqxB overexpression was observed in nine eravacycline-nonsusceptible isolates (range, 35.64–309.02-fold) and 13 eravacycline-heteroresistant isolates (8.42–296.34-fold). The overexpression of macA or macB was detected in 12 eravacycline-heteroresistant isolates (3.23–28.35-fold). Overexpression of the efflux pump regulator gene ramA was observed in 11 eravacycline-nonsusceptible isolates (3.33–94.05-fold) and 18 eravacycline-heteroresistant isolates (3.89–571.70-fold). The eravacycline MICs were increased by one–fourfold by overexpression of oqxAB or macAB in three eravacycline-sensitive isolates. In conclusion, the overexpression of OqxAB and MacAB efflux pumps and the transcriptional regulator RamA were suggested to be involved in K. pneumoniae eravacycline resistance and heteroresistance.
These authors contributed equally: Jin-xin Zheng, Zhi-wei Lin, Xiang Sun, Wei-hong Lin
Introduction
Concerns regarding the Gram-negative pathogen Klebsiella pneumoniae, a member of the Enterobacteriaceae family, are growing worldwide due to the increasing incidence of severe infections, antibiotic-resistant strains, and reduced treatment efficacyCitation1. Carbapenem-resistant Enterobacteriaceae are an emergent global health threat because carbapenems had previously been effective for eliminating multidrug-resistant Gram-negative bacterial infectionsCitation2. In particular, increases in carbapenem-resistant K. pneumoniae (CR-Kp) frequencies worldwide are resulting in K. pneumoniae infections that are very difficult to treat and are thus associated with higher mortality ratesCitation3.
Tigecycline, the original member of the glycylcycline group of antibiotics, has been shown to have antimicrobial activity against CR-Kp in vitro, and thus, this antibiotic may be a last resort therapeutic option against CR-Kp infectionsCitation4. However, cases of tigecycline-nonsusceptible K. pneumoniae (TNSKP) have emerged in hospitals with wide clinical application of tigecyclineCitation5–Citation7. In recent years, TNSKP has been reported to occur in patients without prior exposure to tigecyclineCitation8–Citation10. The mechanisms underlying tigecycline resistance are complex and not yet well understood. The overexpression of the efflux pumps AcrAB and OqxAB has been shown to play a crucial role in tigecycline resistance in K. pneumoniaeCitation9,Citation11. Meanwhile, mutations in ramR, acrR, and rpsJ genes have also been reported to contribute to K. pneumoniae resistance to tigecyclineCitation9,Citation12.
Eravacycline, previously known as TP-434, is a novel fluorocycline antibiotic with broad-spectrum activity against Gram-positive and Gram-negative aerobic and anaerobic pathogens in vitroCitation13. Especially noteworthy is the observation that eravacycline has efficacy against several critical antimicrobial-resistant pathogens, such as methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, extended-spectrum β-lactamase (ESBL), carbapenemase-producing Enterobacteriaceae, and multidrug-resistant Acinetobacter baumanniiCitation14–Citation16. Indeed, eravacycline has been reported to be two–fourfold more effective than tigecycline against common clinical Gram-positive and Gram-negative aerobic bacterial speciesCitation13.
Like tigecycline, eravacycline can overcome most prevalent tetracycline-resistant mechanisms. Notably, Grossman et al. found that eravacycline efficacy was only slightly or undetectably affected by common tetracycline resistance factors, including efflux pumps (tetA, tetB, and tetK) and ribosomal protection protein (tetM) variants in Escherichia coli. Meanwhile, the antibacterial potencies of tigecycline and eravacycline were reduced by 4- to 16-fold in two nonisogenic Propionibacterium acnes isolates harboring a 16S rRNA gene (G1058C) mutation compared with that of the wild-type control strainCitation15. The MIC for eravacycline in adeB-hyperexpressing A. baumannii was shown to be reduced by eightfold by disrupting the gene adeBCitation17. However, the traits and mechanisms of eravacycline resistance among clinical K. pneumoniae isolates, especially TNSKP isolates, have yet to be clarified. Thus, the aim of the present study was to explore the characteristics and mechanisms of eravacycline resistance among clinical K. pneumoniae isolates.
Results
Tigecycline and eravacycline susceptibilities among clinical K. pneumoniae isolates
As shown in Table , tigecycline-nonsusceptible isolates of K. pneumoniae were similarly represented between ESBL-positive and -negative strains, as well as between carbapenem-resistant and -susceptible strains. Eravacycline susceptibility was less common among ESBL-positive strains than among ESBL-negative strains (P < 0.05). Among the ESBL-positive isolates, eravacycline nonsusceptibility was more common than tigecycline nonsusceptibility (21.7% vs. 9.4%, P = 0.001).
Tigecycline and eravacycline susceptibility characteristics among 393 clinical K. pneumoniae isolates
Effects of efflux pump inhibitor (EPI) and ribosomal protein gene mutations on K. pneumoniae MICs
To investigate the mechanisms of eravacycline resistance in K. pneumonia, 37 clinical K. pneumoniae isolates were selected for further study, including the tigecycline- or eravacycline-susceptible or nonsusceptible isolates. To explore the differences between eravacycline and tigecycline resistance, these 37 isolates were divided into three groups, those that had MICs of tigecycline that were <, =, or > the MICs of eravacycline. The effects of the EPI Phe-Arg-β-naphthylamide (PAβN) on tigecycline and eravacycline MICs are reported in Table . Notably, among 25 tigecycline- and/or eravacycline-nonsusceptible isolates (MIC ≥4 mg/L), six isolates showed a 16-fold decrease, 10 showed an eightfold decrease, eight showed a fourfold decrease, and one isolate showed a twofold decrease in tigecycline and/or eravacycline MICs in the presence of PAβN (50 mg/L). However, among 12 tigecycline- and eravacycline-susceptible isolates (MIC ≤ 2 mg/L), three showed a fourfold decrease and six showed a twofold decrease in tigecycline and/or eravacycline MICs in the presence of PAβN (50 mg/L).
Tigecycline and eravacycline MICs in the absence or presence of PAβN, and ribosomal protein gene mutations in 37 clinical K. pneumoniae isolates
Mutations in acrR were observed in seven of the above 37 clinical K. pneumoniae isolates and in five of the 25 tigecycline- and/or eravacycline-nonsusceptible isolates (Table ). Mutations in ramR were detected in five of these 37 clinical K. pneumoniae isolates and in three of the 25 tigecycline- and/or eravacycline-nonsusceptible isolates. Three isolates were observed with both ramR and acrR mutations, two of which were tigecycline- and/or eravacycline-nonsusceptible and had MICs comparable to other nonsusceptible isolates. None of the isolates in this study had rpsJ mutations.
Furthermore, we detected these ribosomal protein gene mutations in 47 tigecycline- and eravacycline-sensitive clinical K. pneumonia isolates. Mutations in acrR or ramR were also observed in these eravacycline-sensitive isolates, but were only detected in four isolates and were different from those mutations detected in eravacycline-nonsusceptible isolates (Table S1).
Expression of efflux pump and regulator genes in clinical K. pneumoniae isolates
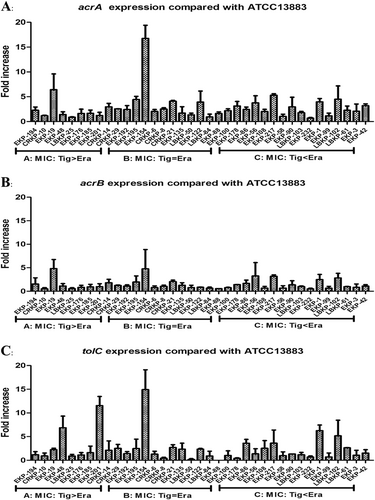
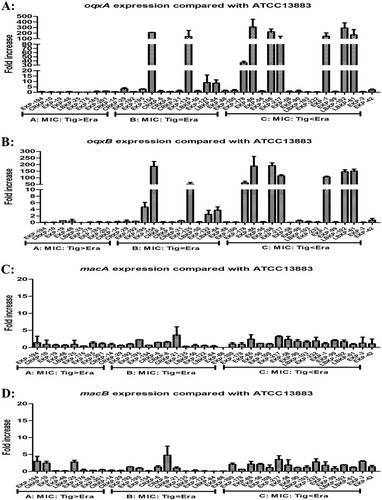
Our qRT-PCR experiments indicated that only 6/37 isolates overexpressed the AcrAB–TolC pathway genes acrA and/or tolC (>fivefold greater than the tigecycline-susceptible K. pneumoniae ATCC 13883 reference strain), with a maximum value detected as 16.77-fold compared with the reference strain (Fig. ). Interestingly, 11/37 isolates were observed to overexpress oqxA or oqxB (range, 8.46–309.02-fold compared with the reference strain levels), with a maximum value of 309.02-fold (Fig. ). Among these 11 isolates, nine exhibited a significant overexpression of oqxAB (35.64–309.02-fold) and all nine isolates were tigecycline- and/or eravacycline-nonsusceptible, with observed eravacycline MICs greater than or equal to the tigecycline MICs in all nine isolates. Two isolates were observed to overexpress acrF (6.55- and 19.74-fold) (Fig. S1). Only 11/37 isolates were observed to overexpress ramA (3.33–94.05-fold), and only 8/37 isolates were observed to overexpress soxS (2.02–11.39-fold) (Fig. S2).
Efflux pump activity and ribosomal protein gene mutations in eravacycline-heteroresistant isolates
A population analysis profile (PAP) of eravacycline susceptibility (MIC <2 mg/L) among clinical K. pneumoniae isolates indicated that 20 isolates could be classified as having heteroresistance to eravacycline. PAβN (50 mg/L) exposure resulted in eravacycline resistance reductions that were modest (four- to eightfold reduction) in 5/20 heteroresistant isolates, moderate (16- to 32-fold) in 7/20 heteroresistant isolates, pronounced (64-fold) in 5/20 heteroresistant isolates, and very pronounced (128-fold) in 3/20 heteroresistant isolates (Table ). These results show that a pharmacological EPI can suppress efflux pump activity in heteroresistant isolates beyond that exhibited in the eravacycline-resistant isolates of K. pneumoniae.
Eravacycline MICs in the absence or presence of PAβN and ribosomal protein gene mutations in 20 eravacycline-heteroresistant clinical K. pneumoniae isolates
Only 3/20 eravacycline-heteroresistant isolates were observed to have acrR mutations, whereas 8/20 eravacycline-heteroresistant isolates were observed to have ramR mutations (Table ). None of the 20 eravacycline-heteroresistant isolates had an rpsJ mutation.
Pump and regulator gene expression in eravacycline-heteroresistant isolates
Among the 20 eravacycline-heteroresistant isolates studied, oqxA (15.61 ± 22.23-fold) or oqxB (22.63 ± 64.90-fold) was significantly overexpressed, relative to the reference strain, in 13/20 eravacycline-heteroresistant isolates (Fig. ). Overexpression of macA (8.26 ± 8.43-fold) or macB (3.44 ± 4.59-fold), relative to the reference strain, was observed in 12/20 eravacycline-heteroresistant isolates (Fig. ). While only seven eravacycline-heteroresistant isolates were observed to overexpress acrA, acrB, or tolC (≥fivefold compared with the reference strain K. pneumoniae ATCC 13883), the mean fold differences in the expression of these genes relative to the reference strain were 4.36 ± 2.84, 3.48 ± 1.72, and 2.91 ± 2.76, respectively (Fig S3). Regarding efflux pump regulator genes, 18/20 eravacycline-heteroresistant isolates were observed to overexpress ramA relative to the reference strain (mean, 147.65 ± 164.40-fold; maximum, 571.70-fold; Fig S4), and the overexpression of ramA in these eravacycline-heteroresistant isolates exceeded that observed in eravacycline-resistant isolates (14.58 ± 30.15-fold). Notably, 20 eravacycline-heteroresistant isolates were observed to include eight isolates with ramR mutations (40.0%), while only 3/23 eravacycline-resistant isolates were observed to have ramR mutations (13.0%) (Tables and ).
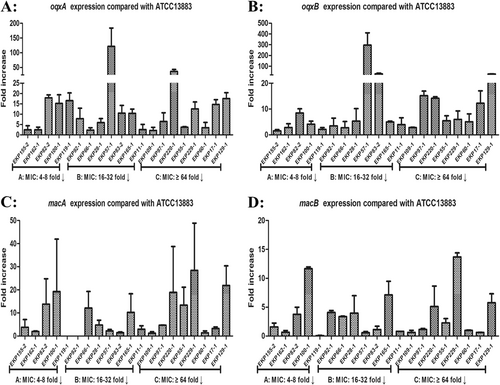
Expression levels were detected by qRT-PCR, with tigecycline-susceptible K. pneumoniae ATCC 13883 used as the reference strain (expression = 1). MIC: 4–8-fold↓, PAβN reduced eravacycline MICs by 4–8-fold compared to eravacycline alone
Mutations in OqxAB and MacAB efflux pumps
Mutations in oqxAB and macAB genes in the assayed clinical K. pneumoniae isolates overexpressing oqxAB or macAB (overexpressed >threefold greater than the ATCC 13883 reference strain) were identified. Mutations in OqxAB and MacAB efflux pumps were rarely reported from previous studies. In this study, only two eravacycline-heteroresistant isolates with oqxAB mutations (EKP82-2 and EKP119-1) and one isolate with a macB mutation (EKP82-2) were identified, which were not present among those eravacycline-resistant isolates (Table S2 and S3).
OqxAB and MacAB efflux pumps associated with eravacycline resistance were tested in an overexpression experiment
To confirm the roles of OqxAB and MacAB efflux pumps in eravacycline resistance of K. pneumoniae, the overexpression of oqxAB and macAB in three eravacycline-sensitive isolates was conducted. As Fig. shows the expression levels of oqxA increased 6.77–7.63-fold and that of macA increased 4.76–5.65-fold, following a 0.2% arabinose (Ara) induction. Interestingly, the eravacycline MICs of these three eravacycline-sensitive isolates increased one–fourfold following the 0.2% Ara induction, relative to controls not exposed to Ara (Table ).
The expression levels of oqxA (a) or macA (b) was determined by qRT-PCR. The wild-type strain of these three isolates were used as the reference strains (expression = 1.0)
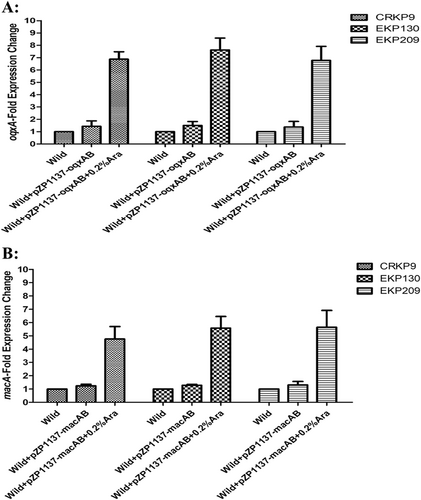
Overexpression of OqxAB or MacAB in three eravacycline-sensitive clinical K. pneumoniae isolates
Discussion
In previous studies, researchers observed that eravacycline had a more powerful antibacterial effect than tigecycline against clinically common aerobic bacterial speciesCitation13,Citation15. However, the results of this study showed that eravacycline-nonsusceptible clinical K. pneumoniae isolates were encountered more frequently than tigecycline-nonsusceptible isolates, especially among ESBL-positive isolates. The occurrence of antibiotic resistance varies regionally. Until now, there have been no reports of eravacycline susceptibility in K. pneumoniae from China. We believe that the K. pneumoniae isolates from different regions provide an explanation for different results, but this issue needs additional study in China for confirmation. In the present study, the overexpression of several antibiotic resistance-related genes, including genes encoding proteins that constitute the resistance-nodulation-cell division (RND)-type efflux pump AcrAB, the quinolone/olaquindox efflux pump OqxAB, and the transcriptional activators RamA and SoxS, was observed among the examined eravacycline-nonsusceptible isolates. Imipenem, meropenem, and colistin heteroresistances have been documented previously in carbapenemase-producing isolates of K. pneumoniaeCitation18–Citation21. To the best of our knowledge, this is the first study to clinically document K. pneumoniae isolates with eravacycline heteroresistance.
The RND-type efflux pump, AcrAB-TolC, has been shown to play an important role in Enterobacteriaceae antimicrobial resistance, especially tigecycline resistance in K. pneumoniaeCitation4,Citation9,Citation22,Citation23. Quinolone/olaquindox efflux pump, OqxAB, was also associated recently with tigecycline resistance in K. pneumoniaeCitation8,Citation11. The overexpression of AcrAB-TolC and OqxAB in clinical K. pneumoniae isolates without a history of tigecycline exposure may be due to exposure to other antimicrobials that are transported by the same efflux pumpsCitation8–Citation12. The eravacycline-resistant and eravacycline-heteroresistant K. pneumoniae isolates assayed in this study had not been exposed to tigecycline or eravacycline, but their overexpression of AcrAB-TolC and/or OqxAB is consistent with the possibility of their exposure to other compounds transported by these pumps. Although the overexpression of both AcrAB-TolC and OqxAB have previously been implicated in tigecycline resistance in K. pneumoniae; in this study, we observed a more pronounced OqxAB overexpression relative to that of AcrAB-TolC in eravacycline-resistant and -heteroresistant isolates of K. pneumoniae. Similar to previous studies, we observed that the MICs of some eravacycline-resistant and -heteroresistant isolates were inhibited significantly by PAβN, despite the low-level expression of AcrAB-TolC or OqxABCitation9. Eravacycline resistance in these K. pneumoniae isolates may be due to efflux pumps other than AcrAB-TolC or OqxAB.
The overexpression of the RND-type efflux pump AcrEF was shown to be associated with fluoroquinolone resistance in Salmonella enterica serovar TyphimuriumCitation24. A recent study by Zhang et al.Citation25 observed that AcrEF upregulation contributes to quinolone resistance development in E. coli. Two eravacycline-resistant K. pneumoniae isolates (EKP195 and EKP56) that overexpressed AcrF in the present study provide the first report of the involvement of AcrF in K. pneumoniae tetracycline resistance.
This study provides the first evidence implicating the overexpression of the periplasmic adapter MacAB in eravacycline resistance and heteroresistance in clinical K. pneumoniae isolates. The MacAB-TolC pump assembly, which is now the best-studied bacterial ABC drug exporter, was demonstrated in E. coli to be a cell envelope-spanning transmembrane transporter that actively extrudes the substrates, including macrolide antibiotics and polypeptide virulence factorsCitation26,Citation27. Our observation of high levels of macA expression in 12 eravacycline-heteroresistant isolates indicates that the MacAB-TolC multidrug efflux pump may, like OqxAB, play an important role in eravacycline heteroresistance in K. pneumoniae. Previous studies have indicated that tigecycline resistance in K. pneumonia is primarily due to the overexpression of AcrAB efflux pump. However, the results of this study showed that the OqxAB and MacAB efflux pumps play a more important role than AcrAB in the development of eravacycline resistance in K. pneumonia. The reason for this difference may be that the C-7 and C-9 of the tetracycline core D-ring of eravacycline are notably different from those in tigecycline, although the two antibacterials belong to the tetracycline familyCitation13.
The expression of AcrAB-TolC efflux pump can be modulated by several transcriptional regulators, including those encoded by ramA, marA, rarA, soxS, and robACitation4,Citation22,Citation23. In the present study, we observed the overexpression of transcriptional regulator ramA in our eravacycline-resistant and -heteroresistant K. pneumoniae isolates without the overexpression of AcrAB-TolC efflux pump-encoding genes. Interestingly, the overexpression of OqxAB and MacAB-TolC efflux pump-encoding genes was observed in eravacycline-resistant and -heteroresistant K. pneumoniae isolates. The efflux pump OqxAB is activated by the transcriptional regulators RamA and RarACitation8,Citation28. The observed overexpression of OqxAB in this study may have been related to the upregulated expression of RamA, rather than being attributable to RarA or other transcriptional regulators. The ramR gene is located upstream of ramA, and ramR is a negative regulator of ramA. Thus, diverse ramR mutations can lead to the upregulation of ramA expression and subsequently contribute to reduced antibiotic susceptibilityCitation9,Citation28. In this study, the eravacycline-heteroresistant isolates with higher ramA expression than those in eravacycline-resistant isolates may be due to the heteroresistant isolates having more ramR mutations. However, among some eravacycline-resistant (EKP195, EKP154, CRKP21, and EKP217) and -heteroresistant (EKP162-1, EKP82-2, EKP100-1, EKP66-1, EKP57-1, EKP55-1, EKP229-1, EKP17-1, and EKP129-1) isolates, we observed elevated ramA expression in the absence of any ramR mutations. Thus, the mechanisms responsible for high ramA expression may due to other unknown transcriptional regulators in those isolates and require further investigation.
Nonetheless, the above-reported efflux pumps cannot explain the observed eravacycline resistance observed in four isolates (EKP192, CRKP6, CRKP21, and EKP56), whose MICs were inhibited significantly by PAβN despite the low-level expression of efflux pump genes. Therefore, the underlying mechanisms for these eravacycline-resistant isolates need to be further studied. The role of the MacAB-TolC multidrug efflux pump in K. pneumoniae resistance to eravacycline also requires further study.
In conclusion, the results of this study implicate the overexpression of OqxAB, but not AcrAB-TolC, as well as the overexpression of the transcriptional regulator RamA in K. pneumoniae eravacycline resistance and heteroresistance. Moreover, the present data suggest that the MacAB-TolC multidrug efflux pump may also participate in eravacycline resistance and heteroresistance in K. pneumoniae.
Materials and methods
Bacterial strains and growth conditions
From January 2010 to December 2016, 393 nonduplicate K. pneumoniae isolates from various clinically sampled infections of individuals not previously exposed to tigecycline or eravacycline were collected from in-patients at Shenzhen Nanshan People’s Hospital at Shenzhen University School of Medicine in China. The strains were identified with a Phoenix 100 automated microbiology system (BD, Franklin Lakes, NJ, USA) and then, two subcultured generations were reidentified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (IVD MALDI Biotyper, Germany). All strains were cultured in Luria-Bertani medium at 37 °C. All procedures involving human participants were performed in accordance with the ethical standards of the Shenzhen University School of Medicine and with the 1964 Helsinki declaration and its later amendments. For this type of study, formal consent is not required.
Antimicrobials
Two carbapenem drugs, namely imipenem (catalog no. MB1457) and meropenem (catalog no. MB1129), were purchased from Meilunbio (Dalian, China). The glycylcycline drug tigecycline (catalog no. E129449) was purchased from Aladdin (Shanghai, China). The novel fluorocycline drug eravacycline (catalog no. A13887-10) was purchased from AdooQ BioScience (Irvine, CA, USA).
Antimicrobial susceptibility testing
The ESBL production of K. pneumoniae isolates was detected using the Phoenix 100 automated microbiology system. MICs for imipenem, meropenem, tigecycline, and eravacycline were determined by the agar dilution method, according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI-M100-S26). The MIC breakpoints for carbapenem were defined according to CLSI-M100-S26. Isolates observed to have elevated MICs for carbapenem were confirmed by a manual ETEST® (bioMérieux) to have a reduced susceptibility to either imipenem or/and meropenemCitation29.
PAP analysis
PAPs were obtained as described previouslyCitation30. One-hundred-microliter aliquots of a starting cell suspension (corresponding to a 0.5 McFarland standard for K. pneumoniae cultures grown on blood agar plates for 24 h at 37 °C; 1.0–1.5 × 108 cfu/ml) were spread onto Mueller–Hinton agar plates with or without eravacycline (1, 2, 4, 6, 8, or 16 mg/l). After incubating for 24 h at 37 °C, the number of colonies was counted. As CLSI Enterobacteriaceae MIC breakpoints for tigecycline and eravacycline have not yet been established, K. pneumoniae isolates with an MIC ≥4 mg/l for tigecycline or eravacycline were considered to be not susceptible based on the reference studiesCitation17,Citation31. Eravacycline heteroresistance was defined as the presence of eravacycline-susceptible isolates with an eravacycline MIC of <2 mg/l, in which detectable subpopulations grew in the presence of ≥4 mg/l eravacycline, with a detection limit of 20 cfu/ml. Each analysis was performed three times.
Efflux inhibition
Efflux pump activity in tigecycline- and/or eravacycline-resistant K. pneumoniae isolates was detected using the efflux pump inhibitor (EPI) Phe-Arg-β-naphthylamide (PAβN, Sigma). Tigecycline and eravacycline MICs were determined by the agar dilution method in the presence and absence of PAβN (50 mg/l). Significant inhibition of efflux pumps was confirmed based on the MIC reduction to a quarter (or more) of the baseline values in the presence of EPICitation8.
qRT-PCR
Transcript expression levels of the efflux pump genes acrA, acrB, tolC, oqxA, oqxB, acrE, acrF, macA, and macB and their transcriptional regulator genes acrR, marA, soxS, rarA, robA, and ramA were determined by qRT-PCR, with the primers listed in Table S4. Total bacterial RNA was extracted with an RNeasy mini kit (QIAGEN GmbH, Hilden, Germany), and cDNA was synthesized with a PrimeScript RT reagent kit (TAKARA BIO INC, Shiga, Japan). Finally, qRT-PCR was performed using a SYBR Premix Ex Taq II kit (TAKARA BIO INC, Shiga, Japan) in a Mastercycler ep realplex system (Eppendorf, Hamburg, Germany), with an initial incubation at 95 °C for 2 min, followed by 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Each sample was run in triplicate. The expression of target genes was normalized relative to the 16S rRNA housekeeping gene rrsE. Threshold cycle (Ct) numbers were confirmed by the qRT-PCR system software, and data were analyzed in accordance with the 2−ΔΔCt method. The expression levels of the target genes were compared with those of K. pneumoniae ATCC 13883 (tigecycline susceptibleCitation9 and eravacycline susceptible [according to our sensitivity test] strain, expression = 1).
Detection of mutations in acrR, ramR, rpsJ, oqxAB, and macAB
The acrR, ramR, and rpsJ codons of the S10 ribosomal protein-encoding genes, as well as those of the efflux pump-encoding genes oqxAB and macAB were amplified by PCR (primers listed in Table S4) and sequenced. Mutations in acrR, ramR, and rpsJ in K. pneumoniae isolates were identified by comparison with a reference sequence, namely, the genome of K. pneumoniae subsp. pneumoniae MGH 78578 (GenBank accession number CP000647)Citation9. Mutations in oqxAB and macAB were identified by comparison with reference sequences from the NCBI database (for detailed sequences see Tables S2 and S3 footnotes).
Overexpression of OqxAB and MacAB in eravacycline-sensitive isolates of K. pneumoniae
Complete oqxAB gene was amplified by PCR from the eravacycline-heteroresistant clinical isolate EKP82-2, and the macAB gene was amplified from eravacycline-heteroresistant clinical isolate EKP129-1. The PCR fragments were purified and digested with endonucleases NheI and BglII and then were inserted into the plasmid pZP1137 for gene overexpression. Correct cloning was verified by PCR and sequencing. Verified plasmid constructs were introduced into three eravacycline-sensitive K. pneumoniae clinical isolates: CRKP9, EKP130, and EKP209. All strains, plasmids, and primers used for overexpression are listed in Tables S5 and S6. The overexpression of OqxAB and MacAB was induced with 0.2% arabinose (Ara). All assays were performed at least in triplicate.
Statistical analysis
Continuous data are reported as the means ± standard deviations (SDs) and were analyzed with Student’s t tests, one-way factorial analyses of variance (ANOVA), or nonparametric Mann–Whitney U tests. Categorical data are reported as numbers (with percentages) and were compared with Chi-square or Fisher’s exact tests. P values <0.05 were regarded as significant. All data were analyzed in SPSS (version 17.0, Chicago, IL, USA).
Supplementary Figure S1
Download TIFF Image (315.4 KB)Supplementary Figure S2
Download TIFF Image (696.3 KB)Supplementary Figure S3
Download TIFF Image (679.1 KB)Supplementary Figure S4
Download TIFF Image (831.1 KB)Supplementary Information
Download MS Word (32.1 KB)Acknowledgements
The authors thank Prof. Yun-song Yu (Department of Infectious Diseases, Sir Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang, China) for generously providing K. pneumoniae ATCC 13883. In addition, we thank Prof. Dao-guo Zhou (Department of Biological Sciences, Purdue University, West Lafayette, Indiana, USA) for generously providing the plasmid pZP1136. We also thank Ms. Cynthia Brast (University of Florida, Gainesville, FL, USA) for her review of this manuscript. This work was supported by grants from the National Natural Science Foundation of China (grant number 81170370); the Sanming Project of Medicine in Shenzhen (grant number SMGC201705029); the Shenzhen Scientific Research Program (grant numbers JCYJ20170412143551332, JCYJ20170307153714512, JCYJ20170307153425389, and JCYJ20170307153919735); the Scientific Research Project of the Shenzhen Health and Family Planning System (grant number 201601058); and the Shenzhen Nanshan District Scientific Research Program of the People’s Republic of China (grant number 2016010).
Authors’ contributions
J.Z. participated in the design of the study, carried out the qRT-PCR assays and interpreted the data, and drafted the manuscript. Z.L., X.S., and W.L. performed antibiotic susceptibility tests, PAP analyses, qRT-PCR assays, detected mutations in ribosomal protein-encoding genes, performed the overexpression of oqxAB and macAB test, and participated in data analysis. Y.W. and Z.C. participated in antibiotic susceptibility tests and efflux inhibition assays. G.Q. participated in qRT-PCR assays and the detection of mutations in ribosomal protein-encoding genes. Q.D., D.Q., and Z.Y. designed the study, participated in data analysis, and provided critical revisions of the manuscript for important intellectual content.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41426-018-0141-y).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- PaczosaMKMecsasJKlebsiella pneumoniae: going on the offense with a strong defenseMicrobiol. Mol. Biol. Rev.201680 629 66110.1128/MMBR.00078-154981674
- ThadenJTPogueJMKayeKSRole of newer and re-emerging older agents in the treatment of infections caused by carbapenem-resistant EnterobacteriaceaeVirulence2017840341610.1080/21505594.2016.1207834
- ZhengBMolecular epidemiology and risk factors of carbapenem-resistant Klebsiella pneumoniae infections in Eastern ChinaFront. Microbiol.20178106110.3389/fmicb.2017.010615468447
- RoySDattaSViswanathanRSinghAKBasuSTigecycline susceptibility in Klebsiella pneumoniae and Escherichia coli causing neonatal septicaemia (2007-10) and role of an efflux pump in tigecycline non-susceptibilityJ. Antimicrob. Chemother.2013681036104210.1093/jac/dks535
- Rodriguez-AvialCRodriguez-AvialIMerinoPPicazoJJKlebsiella pneumoniae: development of a mixed population of carbapenem and tigecycline resistance during antimicrobial therapy in a kidney transplant patientClin. Microbiol. Infect.201318616610.1111/j.1469-0691.2011.03482.x
- TsaiHYEmergence of tigecycline-resistant Klebsiella pneumoniae after tigecycline therapy for complicated urinary tract infection caused by carbapenem-resistant Escherichia coliJ. Infect.20126558458610.1016/j.jinf.2012.09.007
- SpanuTIn vivo emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coliAntimicrob. Agents Chemother.2012564516451810.1128/AAC.00234-123421558
- ZhongXFirst emergence of acrAB and oqxAB mediated tigecycline resistance in clinical isolates of Klebsiella pneumoniae pre-dating the use of tigecycline in a Chinese HospitalPLoS ONE20149e11518510.1371/journal.pone.01151854264890
- LiRLTigecycline susceptibility and molecular resistance mechanisms among clinical Klebsiella pneumoniae strains isolated during non- tigecycline treatmentMicrob. Drug Resist.20172313914610.1089/mdr.2015.0258
- ShengZKEmergence of tigecycline-and carbapenem-nonsusceptible Klebsiella pneumoniae ST11 clone in patients without exposure to tigecyclineJ. Microbiol. Immunol. Infect.20164996296810.1016/j.jmii.2015.10.014
- Bialek-DavenetSDifferential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniaeJ. Antimicrob. Chemother.201570818810.1093/jac/dku340
- FangLStep-wise increase in tigecycline resistance in Klebsiella pneumoniae associated with mutations in ramR, lon and rpsJPLoS ONE201611e016501910.1371/journal.pone.01650195072711
- ZhanelGGReview of eravacycline, a novel fluorocycline antibacterial agentDrugs20167656758810.1007/s40265-016-0545-8
- BassettiMRighiEEravacycline for the treatment of intra-abdominal infectionsExpert. Opin. Investig. Drugs2014231575158410.1517/13543784.2014.965253
- GrossmanTHTarget-and resistance-based mechanistic studies with TP-434, a novel fluorocycline antibioticAntimicrob. Agents Chemother.2012562559256410.1128/AAC.06187-113346605
- SutcliffeJAO’BrienWFyfeCGrossmanTHAntibacterial activity of eravacycline (TP-434), a novel fluorocycline, against hospital and community pathogensAntimicrob. Agents Chemother.2013575548555810.1128/AAC.01288-133811277
- AbdallahMActivity of eravacycline against Enterobacteriaceae and Acinetobacter baumannii, including multidrug-resistant isolates, from New York CityAntimicrob. Agents Chemother.2015591802180510.1128/AAC.04809-144325809
- NodariCSRibeiroVBBarthALImipenem heteroresistance: high prevalence among Enterobacteriaceae Klebsiella pneumoniae carbapenemase producersJ. Med. Microbiol.20156412412610.1099/jmm.0.081869-0
- PournarasSCharacteristics of meropenem heteroresistance in Klebsiella pneumoniae carbapenemase (KPC)-producing clinical isolates of K. PneumoniaeJ. Clin. Microbiol.2010482601260410.1128/JCM.02134-092897536
- MeletisGTzampazESianouETzavarasISofianouDColistin heteroresistance in carbapenemase-producing Klebsiella pneumoniaeJ. Antimicrob. Chemother.20116694694710.1093/jac/dkr007
- BardetLDeciphering heteroresistance to colistin in a Klebsiella pneumoniae isolate from Marseille, FranceAntimicrob. Agents Chemother.201761e003561710.1128/AAC.00356-175444162
- BratuSLandmanDGeorgeASalvaniJQualeJCorrelation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York CityJ. Antimicrob. Chemother.20096427828310.1093/jac/dkp1862707265
- PerezAEffect of transcriptional activators SoxS, RobA, and RamA on expression of multidrug efflux pump AcrAB-TolC in Enterobacter cloacaeAntimicrob. Agents Chemother.2012566256626610.1128/AAC.01085-123497196
- OlliverAValléMChaslus-DanclaECloeckaertAOverexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar Typhimurium DT204 acrB mutants selected with fluoroquinolonesAntimicrob. Agents Chemother.20054928930110.1128/AAC.49.1.289-301.2005538886
- ZhangCZUpregulation of AcrEF in quinolone resistance development in Escherichia coli when AcrAB-TolC function is impairedMicrob. Drug Resist.201724182310.1089/mdr.2016.0207
- KobayashiNNishinoKYamaguchiANovel macrolide-specific ABC-type efflux transporter in Escherichia coliJ. Bacteriol.20011835639564410.1128/JB.183.19.5639-5644.200195455
- FitzpatrickAWPStructure of the MacAB-TolC ABC-type tripartite multidrug efflux pumpNat. Microbiol.201721707010.1038/nmicrobiol.2017.705447821
- LinYTIn vivo evolution of tigecycline-non-susceptible Klebsiella pneumonia strains in patients: relationship between virulence and resistanceInt. J. Antimicrob. Agents20164848549110.1016/j.ijantimicag.2016.07.008
- ChiuSKRoles of ramR and tet(A) mutations in conferring tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae clinical isolatesAntimicrob. Agents Chemother.201761e003911710.1128/AAC.00391-175527587
- HalabyTGenomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreakAntimicrob. Agents Chemother.2016606837684310.1128/AAC.01344-165075093
- MarchaimDMajor variation in MICs of tigecycline in Gram-negative bacilli as a function of testing methodJ. Clin. Microbiol.2014521617162110.1128/JCM.00001-143993642