ABSTRACT
Santalum yasi is a high-value hemiparasitic tree endemic to Fiji, Niue and Tonga. It has been overexploited for its oil-yielding heartwood and is now threatened. Remaining stands lack genetic diversity and are likely to be suffering from inbreeding depression, although the species still has significant genetic diversity overall. We argue that the best way to conserve this species is through an active domestication program that will adequately sample and conserve the genetic base in ex situ and circa situm plantings. The approach to S. yasi tree-breeding can be characterised as a low-input strategy involving the early use of molecular markers for population parameter determination. Long-term success will have strong interdependent links with the conservation of the remaining genetic resources. A strategy based on recurrent selection and breeding for key traits—including heartwood volume and oil yield per year, oil quality and environmental adaptability related to cyclone resistance and the tolerance of pests and diseases—is recommended. The establishment of genetic conservation stands based on collections of the species throughout its natural range in Fiji and Tonga has commenced. Challenges associated with the conservation and domestication of S. yasi are discussed. These include the advanced age required before oil characterisation can be undertaken; the need to assess genotype–host-plant interactions; and the need for comparatively sophisticated equipment and destructive harvesting to carry out oil assessments. Capacity development of professional staff in the Pacific Islands is an additional prerequisite for implementing an effective strategy. Research into the variation and heritability of heartwood formation and oil characteristics, and a better understanding of the breeding biology of S. yasi and geneflow between it and exotic Indian sandalwood (S. album), are high priorities. It will be more than a decade—probably around 20 years—before S. yasi individuals in planned, well-designed trial plantings have sufficient heartwood development to enable oil-trait assessment. Establishment of such trials is an immediate priority. In addition to this long-term activity, we recommend a simple interim strategy that promotes high genetic diversity of seedling-based planting stock. This can be implemented using a combination of gene conservation stands, progeny trials that can be culled to seedling seed orchards, and genetically diverse community-based seed stands. The strategy will both provide a safeguard against the further loss of diversity and promote wide outcrossing. Releasing fragmented populations from inbreeding depression is expected to increase general vigour.
Introduction
The sandalwood genus and Santalum yasi
The sandalwoods (Santalum spp.) are a genus of hemiparasitic trees that, collectively, have a natural range from southern Australia, extending north to Indonesia, Sri Lanka and India and east into the Pacific Ocean, from Hawaii in the north to the Juan Fernández Islands in the south-east (Santalum fernandezianum Phil., now extinct). Many of the c. 16 surviving species have commercially valuable heartwood that can be harvested destructively for essential-oil extraction. Of the Santalum species, Santalum album L. (Indian sandalwood) is generally considered the most commercially valuable. Older trees (more than 25–30 years) of S. album typically provide a good yield of high-quality, santalol-rich oil that can be used in perfumery and incense wood applications. Commercial exploitation has been under way for two centuries, with the result that Santalum is considered one of the most heavily exploited groups of plants across its range (Brennan & Merlin Citation1991).
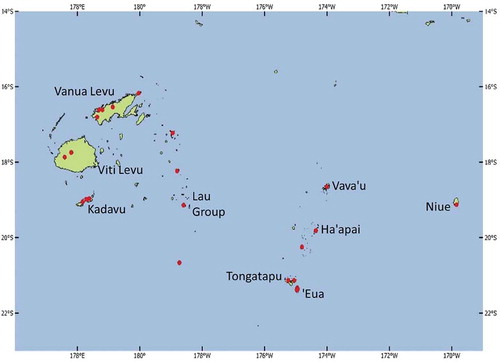
Santalum yasi Seem. is an important cultural and economic plant in Fiji, where it is known as yasi, and in Tonga, where it is called ‘ahi. The natural range extends from Niue and ‘Eua, a southern island in the Tongan group, through Tongatapu, Ha’apai, Vava’u and Niuas in Tonga, and west and north through parts of Fiji—the Lau Group to Bua and Macuata provinces (western Vanua Levu), Udu Peninsula (north-east Vanua Levu), Kadavu and the Nausori Highlands (western Viti Levu) (). The climate in this range is variable, with mean annual temperature (MAT) ranging from 23°C to 29°C and mean annual rainfall (MAR) from 1400 mm to 2500 mm, with either a summer-dominant or bimodal distribution and a dry season of 3–5 months. The natural distribution is mostly lowland up to 300 m above sea level (ASL), but can be as high as 600 m ASL (Elevitch Citation2006).
Figure 1. Natural distribution of Santalum yasi in Fiji and Tonga (after Thomson et al. Citation2018)
Santalum yasi is a small-to-medium (often 6–15 m tall when mature), long-lived tree that often exhibits a multistemmed habit. Thomson et al. (Citation2018) provided a detailed description of the tree’s morphological features. The lower bole may reach diameters of 50 cm after 40–50 years, although trees of this size are now very scarce. A review of historical accounts of exploitation (Huish et al. Citation2015) presents evidence that S. yasi was once abundant in both Fiji and Tonga. However, recent surveys in Fiji and Tonga (Huish et al. Citation2015; Bush et al. Citation2016) have revealed that large mature trees are very rare in the wild, with no extensive, unfragmented wild populations identified in either country. This is clearly attributable to past and ongoing exploitation and regenerative stress.
Although S. yasi is naturally fragmented, with populations on geographically widespread islands and disjunct areas on the larger islands of Vanua Levu and Viti Levu in Fiji, heavy exploitation has caused significant further fragmentation. Individual populations now often comprise only a few trees spread over a wide area of several hectares. Bush and Thomson (Citation2018) considered that the species should be designated as endangered on the International Union for Conservation of Nature (IUCN) Red List. Although those authors did not give detailed reasons for their assessment, the global population would likely meet more than one of the relevant criteria (IUCN Citation2012), any of which, when met, justify a listing, including a reduction in individuals of ≥50% over the last three generations, and criteria related to fragmentation and ongoing exploitation. In Tonga, measures have been taken to control the harvest and export of S. yasi (Motuliki Citation2020), but illegal cutting from the wild and even from plantations and urban gardens still occurs in both Tonga and Fiji.
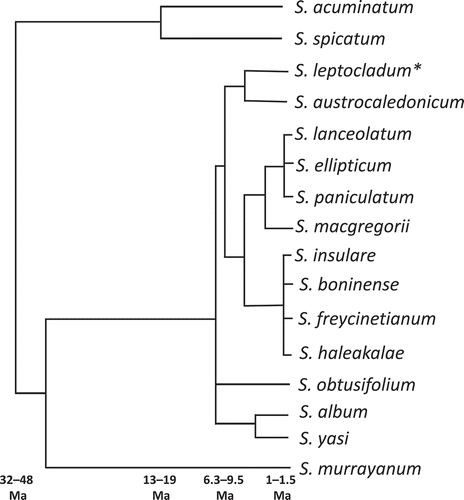
The phylogenetic relationships among the 16 species of the Santalum genus were resolved by Harbaugh and Baldwin (Citation2007) using nuclear ribosomal and strongly conserved chloroplast DNA. Those authors postulated an Australian ancestor, with species radiating out into the Pacific (). According to this phylogeny, S. yasi is most closely related to S. album. Some authors consider that the Tongan population was introduced from Fiji by humans in pre-European times (Brennan & Merlin Citation1991; Harbaugh & Baldwin Citation2007). A study of DNA and morphological characters from a wide range of samples showed that the Fijian and Tongan populations are distinct and genetically differentiated (Bush et al. Citation2016).
Figure 2. Simplified phylogenetic relationships among members of the Santalum genus. Diagram is based on the more-comprehensive chronogram and phylogeny of Harbaugh and Baldwin (Citation2007), which examined multiple populations of each species. The figure incorporates later research (Harbaugh Citation2007) that differentiated the northern subpopulation of S. lanceolatum as S. leptocladum (*)
Cultivation
Santalum yasi grows best in lowland, relatively dry (below about 2000 mm MAR and with a dry season longer than three months), open-forest types in Fiji, Niue and Tonga. It performs well when planted with suitable host plants in home gardens and in smallholder agroforestry and mixed indigenous forest stands. Santalum yasi is very adaptable when it comes to host plants (Elevitch Citation2006; Thomson et al. Citation2018), but the following are particularly good hosts: nitrogen-fixing trees such as Acacia leptocarpa Benth., Acacia richii A.Gray, Cajanus cajan (L.) Millsp., Calliandra calothyrsus Meisn. and Casuarina equisetifolia L.; and other small-to-medium-sized trees such as Citrus maxima (Burm.) Merr., Citrus reticulata Blanco, Citrus × taitensis and Flueggea flexuosa Müll.Arg. In suitable growing conditions, S. yasi may attain harvestable size in about 25 years (20–25 cm diameter at base with substantial heartwood development). It appears that, like Santalum austrocaledonicum Vieill. (Page et al. Citation2012), S. yasi grows at a rate of around 1 cm of basal diameter per year (on average) throughout Fiji and Tonga (Bush, Thomson et al. Citation2020).
Government departments in both Fiji and Tonga advise growers to delay harvesting sandalwood until at least 20 years. There is often a temptation or necessity to generate earlier cash income, however, and trees are commonly harvested at around 15 years, at which time only a modest amount of heartwood has usually developed. A survey of trees in Fiji and Tonga in 2018 indicated that harvesting at this age risks losing substantial income, with incomplete heartwood defvelopment evident (Bush, Brophy et al. Citation2020); similar findings were made by Doran et al. (Citation2005), who examined young-aged (10–20-year-old) sandalwood including S. album, S. yasi and their hybrid in Fiji and Tonga, and by Jones et al. (Citation2006), who studied 14–17-year-old planted S. album at Kununurra, Western Australia.
Diversity and its importance
The genetic diversity of S. yasi has been assessed throughout its range in Fiji and Tonga using microsatellite markers (Bush et al. Citation2016). Due to heavy harvesting, the species is now restricted to fragmented stands in the wild. Nevertheless, the overall population still has moderate genetic diversity, now mainly comprising planted stands and individual trees in many villages and even urban areas. There is significant population differentiation between the Fijian and Tongan populations (overall FST = 11%), with some differences between Kadavu, Vanua Levu and Viti Levu, while the populations of the Tongan Island groups (‘Eua, Ha’apai, Tongatapu and Vava’u) appear very similar to each other. Samples taken from Fiji’s southern Lau island group indicate that the population there has a closer affinity to the Tongan population than to those in Fiji.
Although diversity is moderate, it appears that inbreeding is a problem in all the sampled populations in Fiji and Tonga (Bush et al. Citation2016). This is probably due to the fragmentation of the wild stands and the propagation of planting stock using seeds drawn from a narrow genetic base. Although the impact of inbreeding has not been studied in Santalum species, it has caused strongly deleterious effects, referred to as inbreeding depression, in most tree species where it has been examined (e.g. Sedgley & Griffin Citation1989; Williams & Savolainen Citation1996; Griffin et al. Citation2019). Inbreeding depression in trees typically results in poor seed set, high seedling mortality, the ongoing premature mortality of established trees and a marked reduction in vigour. If inbreeding depression is indeed present, the establishment of broadly based S. yasi seed orchards could result in significant productivity gains. The assessment of provenance-level growth, adaptation and wood properties is also a high priority.
The expression of intraspecific diversity in economically important traits has yet to be comprehensively studied in S. yasi by means of field trials. This is due in part to the difficulty of collecting sufficient quantities of seeds from populations that have been severely depleted by two centuries of heavy exploitation and the lack of a coordinated long-term research program. Nevertheless, it is evident from the species’ natural distribution that different populations have become adapted to widely differing environmental conditions, with MAT and MAR ranges of around 6°C and 1000 mm, respectively. There is considerable variation in heartwood development and oil content between individuals growing in the wild, but well-designed field trials will be required to elucidate the relative importance of genotype, age, environment and interactions for heartwood development. An existing valuable resource is a gene-bank planting at Vunimaqo, Viti Levu, Fiji, which was established by the Australian Agency for International Development (AusAID) CSIRO Pacific Regional Initiative on Forest Genetic Resources (SPRIG) program and the Fiji Ministry of Forestry in 2002; it is a good example of a well-executed gene-bank planting on a secure site.
Mahesh et al. (Citation2018) published a genome for S. album, indicating that this species has the smallest genome of any tree species studied to date (221 mega base), with a somatic chromosome number 2n = 20. Synteny between S. album and other Santalum species is yet to be examined. If S. yasi also has a small genome, this will increase the relative ease of its marker-assisted breeding (see reviews by White et al. Citation2007 and Grattapaglia et al. Citation2018). Marker-assisted breeding involves the association of certain parts of a genome with particular traits and the use of DNA-based information to make selections; it has been problematic for many forest trees because of their large genome sizes and high genetic diversity. Marker-assisted breeding is an advanced breeding option that requires considerable funding and expertise, and it is probably not the most cost-effective or practical short-term option for Pacific sandalwoods. The resolution of relatedness among trees (i.e. definition of a pedigree) in conservation and breeding populations using molecular markers may, however, be useful and achievable.
Based on studies of other Santalum species, S. yasi is likely to have a mixed mating system; that is, it produces a mix of self-pollinated and outcrossed offspring. Some studies (e.g. Ma et al. Citation2006; Tamla et al. Citation2012) suggest that some Santalum species, including S. album, may be facultatively allogamous (i.e. most trees are self-incompatible). Other studies, however, some including S. album, suggest obligate outcrossing (Jyothi et al. Citation1991; Bhaskar Citation1992; Rugkhla et al. Citation1997). If S. yasi is facultatively allogamous, the generally elevated subpopulation-level inbreeding observed by Bush et al. (Citation2016) implies that the species is tolerant of mating between close relatives and/or that it is more than usually tolerant of inbreeding, allowing homozygous alleles to accumulate over successive generations.
Traditionally, early generations of low-input breeding strategies have proceeded under the assumption that, if present, inbreeding may be reasonably homogeneous among families. The application of molecular markers to all 119 open-pollinated families in a first-generation (F1) breeding population of Eucalyptus cladocalyx F.Muell. demonstrated, however, that this assumption can be far from correct, with families varying between completely selfed and completely outcrossed (Bush & Thumma Citation2013). Like most eucalypts, E. cladocalyx has a mixed mating system, but with some self-incompatible individuals (Ellis & Sedgley Citation1992). This situation may be analogous to Santalum species, as indicated by a marker-based study of six families (210 progeny) of Santalum spicatum (R.Br.) A.DC., which were shown to range from predominantly outcrossed to mostly selfed (Muir et al. Citation2007). As a general approach, molecular-marker-based assessments of breeding systems may now be more cost-effective than carrying out controlled pollination studies to assess a breeding system and may now be affordable, even for low-input breeding programs. Although the mechanisms of potential breeding barriers cannot necessarily be inferred from marker-based studies, it is possible to screen far more plants, increasing the likelihood that findings will accurately characterise the breeding population. It is important to gain a better understanding of the breeding system of S. yasi, both for the management of seed production and conservation populations and so that quantitative genetic parameter estimates can be calculated using realistic relationships among progeny.
Hybrids
Santalum yasi hybridises spontaneously with S. album when they are planted together and is likely to hybridise with other Santalum species, although formal studies involving reciprocal crosses are yet to be conducted. Santalum album × S. yasi hybrids have the reputation for rapid growth relative to pure S. yasi, at least at some sites, potentially making their planting a more attractive commercial option. The magnitude of this growth differential has not been formally assessed. More important than mean annual increment of diameter, however, is heartwood production. The relationship between diameter growth and heartwood production at around 30 cm above ground is weak in S. yasi, S. album and their hybrids at ages between about ten and 20 years (Bush, Brophy et al. Citation2020), and a recent study made a similar finding for Santalum macgregorii F.Muell. (Page, Jeffrey et al. Citation2020). The quality of essential oil extracted from the heartwood of hybrids is unknown. First-generation hybrids are likely to produce oil with qualities intermediate to the parents, and indications from small samples of younger-aged trees are that such oil will likely meet the International Organization for Standardization (ISO) standard for East Indian Sandalwood oil (Doran et al. Citation2005; Bush, Brophy et al. Citation2020). Bush et al. (Citation2016) pointed out that inbreeding may be a problem for S. yasi throughout its range, but F1 hybrids are outcrossed. It is possible that the growth differential between the hybrid and S. yasi is at least partly attributable to inbreeding depression of the pure species. The production of outcrossed seed of the pure S. yasi may therefore result in a significant improvement of vigour.
Hybrid breeding is not planned in the next decade, mainly because of a desire to maintain the natural S. yasi gene pool to the greatest extent possible. Another reason is the development of a sandalwood product that is differentiated in the market from S. album. This is possible because the S. yasi oil profile is subtly different to that of S. album and because provenance (i.e. the ‘exotic’ Pacific islands) is important for marketing to end users (Thomson et al. Citation2020).
Propagation
Santalum yasi is commonly propagated by seed, and this is the main method of mass deployment. The species can also be propagated successfully from cuttings and grafting; these methods are likely to be more useful in the development of breeding facilities than for mass propagation, although there is some evidence that clonal S. album can develop heartwood more rapidly than that produced from seed (McComb Citation2009). Cuttings-based propagation may be useful in the future if clones with specific beneficial properties can be selected.
Host-plant interactions
Like all other sandalwood species, S. yasi establishes root connections with one or more host plants via root haustoria (Ouyang et al. Citation2016; Rocha et al. Citation2017). The growth and survival of individual sandalwood trees is strongly dependent on the suitability, arrangement and vigour of the host (Radomiljac et al. Citation1999; Brand Citation2009; da Silva et al. Citation2016). It is important that the size and growth rate of the host is appropriate to the life stage of the sandalwood tree: hosts that are insufficiently large will not support good growth, and hosts that are too vigorous can outcompete and overshadow the sandalwood. For this reason, sandalwood cultivation often involves a series of hosts, including a pot host at the seedling stage and a smaller host at planting. The pruning of very vigorous hosts is another option (see Page et al. Citation2012). The requirement of a host plant presents a special challenge for genetic improvement programs because, in addition to the usual effect of site–genotype interactions on genetic parameter estimation, yasi-genotype–host-genotype interactions need to be taken into account. This, in turn, makes it more complicated to account for site–genotype interactions, especially if different hosts are used or required at different sites.
For the establishment of new trials, it is best to minimise genotype–genotype interactions by using a set of hosts that is consistent within and between trials. Ideally, hosts should be clonal to minimise their genetic diversity, thereby ensuring that each S. yasi genotype is interacting with a consistent set of host genotypes. The common pot host Alternanthera nana R.Br. is ideal because it is easily propagated vegetatively. A wide range of intermediate- and long-term hosts that can also be vegetatively propagated are suitable for S. yasi (see Page et al. Citation2012; Thomson et al. Citation2018).
Wood and essential-oil characteristics
As for other sandalwoods, the three major uses of S. yasi are carvings, incense production and sandalwood oil, all of which are highly valued in Fiji and Tonga for therapeutic and religious purposes. In Tonga, the oil is used to scent tapa cloth and anoint corpses in royal funerals, and S. yasi is also featured in Tongan legends and songs. In Fiji, the oil is used to scent coconut oil and for marriage ceremonies (Thomson Citation2006).
Essential oil extracted from S. yasi heartwood from the lower stem and main roots contains a santalol-rich essential oil. The largest study of S. yasi to date (Bush, Brophy et al. Citation2020) focused on younger-aged trees, as did the previous study of Doran et al. (Citation2005). Both these studies indicated that S. yasi tends to have an oil profile that is rich in β-santalol relative to the ISO standard for S. album (ISO Citation2002), with a significant proportion of trees exceeding the upper limit for this compound. β-santalol is a highly desirable oil constituent considered chiefly responsible for the characteristic sandalwood odour (Baldovini et al. Citation2011); it is argued, therefore, that it would be beneficial to develop a new standard for S. yasi, as is being done for S. austrocaledonicum (Dowell Citation2020), rather than trying to alter the ratios of santalols through selection and breeding.
The heritability of sandalwood oil characters has not been assessed in any species to date due to a lack of suitable common-garden trials that are old enough for the determination to be made. It seems probable that the heritability of oil traits will be moderate to high based on studies of genetic variability of plant-chemistry traits and heartwood characters in other tree species (Bush et al. Citation2011; Santos et al. Citation2016; Mazanec et al. Citation2017). It is also likely that genotype–environment interaction is lower for wood-chemistry traits than for the lower-heritability growth traits (White et al. Citation2007, p. 140). This means that the rate of gain per generation through selection and breeding for increased santalols, altered santalol ratios and/or specific minor components may be greater than for growth traits.
Marker-assisted breeding may also be effective if the chemical pathways associated with oil synthesis are under the control of a relatively small number of genes. The biosynthetic pathways of santalols, santalenes and other S. album oil components have been investigated and some of the responsible genes identified (Jones et al. Citation2006; Jones et al. Citation2008), but their genetic control is not yet fully understood.
Overall objective of the strategy
Securing the future of S. yasi throughout its natural range is important for biodiversity conservation and also because it is a plant of considerable cultural value and commercial potential. To underpin a strategy to conserve and develop the species, basic information on its genetic structure and diversity is required, as well as an assessment of the impact of its hybridisation with introduced species.
To achieve the vision of a sustainable, plantation-based industry, Fiji and Tonga must transition from dependence on wild harvesting to an efficient, plantation-based system, with integrated processing and value-adding (Thomson et al. Citation2020). Significant volumes of S. album oil from northern Australian plantations will progressively enter the market (Thomson Citation2020), so it may be necessary to differentiate S. yasi products. To do this, the remaining S. yasi diversity needs to be secured and the quality of germplasm and plants improved. Recent research (Bush et al. Citation2016) has identified inbreeding in many of the existing stands, which may diminish plantation performance.
The short-term goal for S. yasi is to secure the remaining genetic diversity in Fiji and Tonga. This should be done by establishing ex situ and circa situm conservation stands that collectively have an adequate sample of the remaining diversity of the species. This could be achieved by both collecting seeds and vegetatively propagating selections.
A medium-term goal is to establish provenance-progeny trials at a range of sites in Fiji and Tonga comprising selections from across the species’s native range. The purpose of the trials would be to make growth-based selections for further breeding. In the long term (20+ years), the trials can be used to assess mature heartwood and oil characteristics.
A medium- to long-term goal is to produce genetically improved trees that will maximise the profitability of plantations and downstream industry through enhanced heartwood and heartwood-oil yields and quality.
Breeding strategy overview
SPRIG took initial steps for the breeding of S. yasi with the establishment of gene banks in Fiji and Tonga. These were established in the early 2000s, but limited further progress was made in the following decade, although the popularity of cultivating sandalwood increased. The breeding program recommenced in 2015 with the widespread sampling of trees from wild and planted populations in Fiji and Tonga. The genetic diversity of these populations was assessed using simple sequence repeats (SSR) molecular markers, an unusual early step that yielded excellent information on population structure, diversity and inbreeding that has since guided conservation and breeding-strategy development. The strategy should be low-input sensu Lindgren (Citation2003), with recurrent selection for general combining ability (RS-GCA), which is the strategy used in most forest tree genetic improvement programs (White et al. Citation2007). A low-input strategy is appropriate given the financial and human resources likely to be available and given the scale of planting, which is expected to increase steadily if it is to underpin industry development (Thomson et al. Citation2020) in the next two decades.
As an ongoing recurrent process, a breeding strategy accumulates benefits over successive generations of a cycle of testing, selection and mating (). In RS-GCA strategies, genetic testing follows selection and the selections are ranked for the target traits, with poor performers excluded from the next generation of mating programs (White et al. Citation2007). Typically, an effective breeding strategy involves the maintenance of a hierarchy of three major types of population (the base, breeding and propagation populations) that can continue to meet the demand for genetically improved planting stock for a fourth population—that is, wood-producing plantations (Harwood et al. Citation2001). In this strategy for a threatened species, however, we also consider a fifth population, the conservation population. The five populations are further outlined below.
Figure 3. Diagram representing recurrent selection and breeding for general combining ability (after Harwood et al. Citation2001)
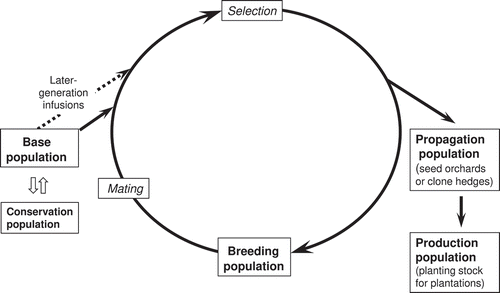
Base population
The base or gene resource population consists of the natural forests of S. yasi in Fiji and Tonga and some plantations in which selection can be carried out. These broad-based reserves will continue to be the source of a wide range of genetic variation to meet future needs.
Conservation population
In the case of S. yasi, it is critically important to take active steps to conserve the remaining diversity in the base population. Under the strategy, conservation plantings will be established comprising unselected samples of the base population sufficient to adequately represent its genetic diversity. The main conservation population can be established as ex situ gene conservation plantings that include a wide range of genotypes comprising an actively managed subpopulation within the base population. At least 60 families as well as grafted trees will be needed to adequately represent the populations present in Fiji and Tonga. This should be expanded over time towards a target of 200 families, with genetic diversity checked using molecular markers. Selection for specific traits will not be practised in this population. In addition, circa situm plantings, the primary purpose of which is not necessarily conservation (e.g. wood production) but which are genetically diverse and situated close enough to existing wild plantings to enable geneflow (via pollen) (Boshier et al. Citation2004; Dawson et al. Citation2013), could be used to complement the role of ex situ plantings as a second conservation population. These circa situm plantings may be genetically improved or include selections for specific traits.
Breeding population
This population comprises selected trees and their progeny in a series of progeny trials and possibly clonal archives, in which the breeding cycle of selection and mating will be repeated over many generations. This is the tree-breeder’s main area of work. An appropriate initial size of this population is about 60 unrelated, open-pollinated families, but ideally this will be expanded over time to upwards of 100 families (a target used, for example, in a series of low-input breeding strategies for newly domesticated eucalypt species in Australia; Harwood et al. Citation2007).
Propagation population
This population consists of intensively selected trees (in this case, initially around 30 selected trees) propagated in seed orchards or cuttings multiplication areas, for which the combinations of genes selected in the breeding population are mass-produced as genetically improved planting stock.
Production population
This population constitutes the major plantation areas established using the improved germplasm.
Infusions of additional material are brought into the breeding population in the second and later generations from the base population to maintain genetic diversity, which is narrowed in each generation through the process of selection. The infusions counteract the accumulation of inbreeding. In the likely case that the initial, F1 breeding population is small (significantly fewer than 100 families), the active acquisition of infusion material will be important to provide adequate diversity for selection and to avoid the accumulation of inbreeding effects.
Selection and mating
Selection and mating are major recurring activities in tree breeding. Genes that influence yield and adaptation are accumulated over successive generations, allowing the breeder to improve desirable traits if they are heritable. The key to an effective breeding strategy is to employ efficient methods for the selection of superior trees. In a low-input strategy, this involves progeny tests at a range of sites in which the selection is carried out. Appropriate measurement techniques and statistical analysis, and selection technologies such as selection indices to integrate the data for several traits (e.g. growth rate, oil yield and oil quality), are required. Mating can be by open pollination or controlled pollination, with care taken to minimise the potential for inbreeding, which may lead to inbreeding depression. The risk of inbreeding can be minimised by designing orchards to reduce the proximity of close relatives, by promoting abundant flowering and by observing flowering intensity to ensure that seed is not collected from genotypes that flower asynchronously. Note that this recommendation is based on the assumption that S. yasi will suffer from inbreeding depression; confirming whether this is the case is a high priority. In the first instance, open pollination is proposed for S. yasi because it is technically easier to manage.
Target planting areas and anticipated scale of planting
The target planting areas may be defined in general terms as suitable site types for the species in Fiji and Tonga. Santalum yasi is intolerant of cool and/or wet soil conditions, precluding some wetter, upland and inland areas on the large islands of Fiji. The species is well adapted to most smaller islands in Fiji and Tonga (Thomson Citation2006; Thomson et al. Citation2018).
The current rate of planting is unknown, although planting at a small scale is popular among farmers, families and villages and encouraged by government agencies in both Fiji and Tonga. A formal assessment of the extent and rate of planting of pure S. yasi and the hybrid in Fiji and Tonga is a high priority.
Selection criteria and traits for selection
The ideal S. yasi tree would be well adapted to target planting environments (displaying excellent survival and health and resistance to attack by pests and diseases); have good vigour, as expressed by rapid stem volume growth with a high proportion of heartwood; have a high oil yield per kilogram of heartwood of preferred santalol compounds; have a straight single bole; and have good resistance to strong winds.
Selection traits for the F1 are usable stem volume; high heartwood proportion (measured by non-destructive coring of living trees); high oil quality; high oil yield per kilogram of heartwood; and resistance to pests and diseases. For selection to be effective and lead to genetic gain, these traits must be heritable in the breeding population. Based on experience with other tree species, tree growth is expected to have low to moderate heritability but the wood and oil traits may be moderately to highly heritable. However, this has not been assessed in any sandalwood species to date.
There are significant challenges for selection in the F1, including a lack of information on heartwood formation relative to growth and a lack of trees of known age. A secondary but still major challenge is a lack of information on genotype–environment interactions with respect to these traits. For example, it is unknown whether climate, soils and cultivation practices act singly or together to influence heartwood properties and oil yield. This challenge is not unique to S. yasi, applying to all sandalwood species. The effectiveness of oil yield and quality analysis from ‘microsamples’ consisting of a few milligrams of wood taken from small-diameter cores also needs further investigation (Bush, Brophy et al. Citation2020). Obtaining non-destructive heartwood samples from trees that are representative of their potential yield will be crucial for the genetic improvement of heartwood and oil traits.
Genotype–environment and genotype–genotype interactions
An initial priority is the establishment of trials across a range of environments involving a wide range of S. yasi genotypes. As a first stage, trials of unselected individuals from across the natural range should be established at as many sites in Fiji and Tonga as practical.
A challenge particular to sandalwood species is the genotype–genotype interaction between the species and its host. To our knowledge, this phenomenon has not previously been studied systematically in any other tree species. Genotype–host interactions should be studied using a restricted range of host genotypes that are not themselves highly genetically diverse to minimise trial size and analytical complexity. The assessment of optimal hosts for planting sandalwood in different situations should be addressed separately. An appropriate model for this research is to use a narrow range of sandalwood genotypes to test various hosts, an approach that has been attempted with S. album tissue-cultured clones and three host species in Western Australia (McComb Citation2009).
Personnel and funding
The availability of technical expertise and institutional support, and an appropriate level of long-term funding, are key elements in determining the type of breeding strategy adopted. Capacity development among technical and research staff is a major priority in both Fiji and Tonga. This may be the most important prerequisite for achieving a long-term vision for sandalwood development. At present, only a small number of staff in the two nations has the required background and technical competencies to carry out some of the more technically challenging aspects of the program. The program requires applied skills in areas such as plus-tree selection and capture, propagation, seed-orchard management, and data management, as well as advanced skills in wood processing, wood chemistry and quantitative and population genetics. Capacity-development activities to support tree improvement and associated research and development should be high priorities in the short, medium and long terms.
Challenges to be addressed
Several challenges need to be addressed to realise the vision of a plantation-based S. yasi industry using genetically improved material in Fiji and Tonga. The ongoing harvesting of wild material and associated loss of genetic diversity is a challenge common to a number of commercially valuable and overexploited Pacific trees (Bush & Thomson Citation2018). However, there are also examples of successful programs for threatened Pacific Island tree species, including S. austrocaledonicum in Vanuatu (Page, Doran et al. Citation2020) and Vanuatu whitewood (Doran et al. Citation2012; Page et al. Citation2017) that have captured genetic resources and commenced domestication. Crucially, such programs require ongoing maintenance and support to ensure success.
A further significant challenge in both Fiji and Tonga is locating secure sites with suitable tenure and ownership arrangements. This is particularly important for long-term gene conservation stands. Initially, plantings are being made at government field stations and, in one case, at a training centre run by a religious institute. Ideally, sites should have stable, long-term tenure so that trees are not harvested when ownership changes, and they must be secure to ensure that valuable genetic resources are not harvested illegally.
Santalum yasi often exhibits sporadic flowering and seed set. Although the seed is not recalcitrant, its storage life is limited. It may therefore be impossible to collect seed from a satisfactory number of genotypes in a single year and, because seed cannot be reliably accumulated in storage, the establishment of single-age trials may be difficult to achieve. Cutting and grafting, although possible, are not widely practised, and success rates may be comparatively low. The development of cutting and grafting skills, improved nursery practice and facilities, and further research into cutting and grafting would be beneficial, especially for the capture of trees that rarely or never set seed. Captured clones can be used to establish clonal seed orchards, the manipulation of which, using cultural or chemical flowering stimulants, may eventuate in seed crops.
Actions for the next decade
The decade to 2030 will be crucial for the conservation of sandalwood in Fiji and Tonga, with the opportunity to both secure the genetic resources of the species and advance domestication programs. The main emphasis should be on ensuring that wild genetic resources are captured and secured in gene banks, seed orchards and other plantings. In addition to formal conservation and breeding-program plantings (gene banks, seed orchards), the production of genetically diverse seed crops for deployment to growers will form the backbone of the strategy. Genetically diverse, outcrossed seed should overcome the potential problems of inbreeding and help conserve the S. yasi gene pool.
It will also be important to lay the foundations for more-ssophisticated domestication and breeding activities. Because of the long lead times associated with sandalwood heartwood development, many of the activities associated with estimating genetic parameters for growth, heartwood and oil traits will occur in the decade after next (i.e. 2031–2040). Key activities in the next ten years include the establishment of ex situ conservation plantings from seed, the assembly of gene banks on secure sites by cutting and grafting, and the establishment of genetically diverse seed orchards with seedlings from multiple seed sources in Fiji and Tonga. To enable the future estimation of genetic parameters, trials should be established across a range of sites that include provenance and family accessions. These should be established using a restricted set of genetically uniform, best-bet hosts to minimise sandalwood–host interactions. Separate trials using a restricted set of sandalwood genotypes and a range of prospective hosts should also be established to determine the optimal hosts for particular site types and planting configurations. Further investigation into the effects of inbreeding and inbreeding depression should be made as a high priority because this information on the breeding system will affect the management of conservation, breeding and production populations.
Germplasm should be exchanged between Fiji and Tonga to increase genetic diversity in both countries and provide reciprocal ex situ conservation. The significant population differentiation between Fijian and Tongan subpopulations suggests that this may be worthwhile. Although an argument could be made that the populations in each country should be kept intact by isolation from one another, the ongoing fragmentation and dwindling of wild resources in both countries probably make this impractical. The introgression of genes (or ‘genetic pollution’) into local wild populations is likely to be a smaller problem from exogenous S. yasi than from S. album, which is a common species in both countries.
Obtaining a better understanding of the variation and development of heartwood and oil properties of S. yasi is a key objective for the next decade. Given that suitable common-garden trials are not yet available, other approaches for collecting data are required. The analysis of wood and oil properties by sampling trees felled for commercial purposes would be highly useful, especially if data on their approximate age, growing environment and hosts at harvest can be gathered. Further non-destructive sampling of planted trees should also be carried out. Gathering such data may provide a better understanding of heartwood development and variation within trees over time. The calculation of the heritability of oil characters is dependent on trial establishment and will probably not be possible until the following decade (2031–2040).
Further developing human capacity and cooperation between Fiji and Tonga will be important in the decade to 2030. There will also be considerable scope for coordinating and harmonising breeding strategy approaches with other Pacific sandalwood species. Santalum album is being commercially developed and genetically improved in Australia (Robson & Barnes Citation2020), and S. austrocaledonicum is being developed in Vanuatu (Page, Doran et al. Citation2020). The tree-breeding program should focus on traits that optimise the development of a viable and equitable value chain that will position S. yasi as a high-value segment of the sandalwood market (Thomson et al. Citation2020); for example, it may be advantageous to deliberately aim for a breed that is rich in β-santalol.
The breeding strategy should be reviewed after ten years (i.e. in 2030). By this time, it is hoped that the S. yasi value chain will have developed and economic breeding objectives can be better defined. Assuming that trials have been established successfully at secure sites, it will be possible to obtain growth data in the ensuing decade and, towards the end of that decade, the collection of wood and oil data. Clearly, the strategy requires a long-term commitment to be successful.
Conclusion
The genetic improvement of S. yasi is a high priority as Fiji and Tonga transition from wild-harvesting to plantation-based production. The conservation and management of existing genetic resources are at least as high a priority as the development of a genetically improved breed of the species. Genetic improvement of tree species is always a long-term undertaking, and this is especially the case for S. yasi due to its slow growth rate and the long period taken to develop good-quality heartwood. Further challenges include the fact that the species is hemiparasitic and the need to take host–genotype interactions into account, in addition to considerations such as genotype–environment interactions. Capacity development among the scientific staff who will manage and oversee the conservation and domestication programs is also a high priority.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Baldovini N, Delasalle C, Joulain D. 2011. Phytochemistry of the heartwood from fragrant Santalum species: a review. Flavour and Fragrance Journal. 26:7–26. doi:10.1002/ffj.2025.
- Bhaskar V. 1992. Pollination biology and fertilization in Santalum album L. (Santalaceae). Flora. 187:73–78. doi:10.1016/S0367-2530(17)32207-7.
- Boshier DH, Gordon JE, Barrance AJ. 2004. Prospects for circa situm tree conservation in Mesoamerican dry-forest agro-ecosystems. In: Frankie GW, Mata A, Vinson SB, editors. Biodiversity conservation in Costa Rica: learning the lessons in a seasonal dry forest. Berkeley (CA): University of California Press; p. 352.
- Brand JE. 2009. Effect of different Acacia acuminata variants as hosts on performance of sandalwood (Santalum spicatum) in the northern and eastern Wheatbelt, Western Australia. Australian Forestry. 72:149–156. doi:10.1080/00049158.2009.10676297.
- Brennan P, Merlin M. 1991. Biogeography and traditional use of Santalum in the Pacific Region. In: McKinnell FH, editor. Sandalwood in the Pacific Region. Australian Center for International Agricultural Research Proceedings. Canberra (Australia): ACIAR; p. 30–39.
- Bush D, Brophy J, Bolatolu W, Dutt S, Hamani S, Doran J, Thomson L. 2020. Oil yield and composition of young Santalum yasi in Fiji and Tonga. Australian Forestry. 83(4):238–244.
- Bush D, McCarthy K, Meder R. 2011. Genetic variation of natural durability traits in Eucalyptus cladocalyx (sugar gum). Annals of Forest Science. 68:1057–1066. doi:10.1007/s13595-011-0121-z.
- Bush D, Thomson L. 2018. Conserving and making efficient use of forest genetic resources. In: Thomson L, Doran J, Clarke B, editors. Trees for life in Oceania. Canberra (Australia): Australian Centre for International Agricultural Research; p. 15–22.
- Bush D, Thomson L, Broadhurst L, Dutt S, Bulai P, Faka’osi T, Havea M, Napa’a S, Vainikolo L. 2016. Assessing genetic diversity of natural and hybrid populations of Santalum yasi in Fiji and Tonga. Canberra (Australia): Australian Centre for International Agricultural Research.
- Bush D, Thomson L, Dutt S, Hamani S, Bolatolu W, Tauraga J, Mateboto J, Young E. 2020. Conservation, domestication and breeding of Santalum yasi in Fiji and Tonga. In: Page T, editor. Sandalwood Regional Forum; 2019 Nov 11–13; Canberra (Australia): Australian Centre for International Agricultural Research.
- Bush D, Thumma B. 2013. Characterising a Eucalyptus cladocalyx breeding population using SNP markers. Tree Genetics & Genomes. 9:741–752. doi:10.1007/s11295-012-0589-1.
- da Silva J, Kher M, Soner D, Page T, Zhang X, Nataraj M, Ma G. 2016. Sandalwood: basic biology, tissue culture, and genetic transformation. Planta. 243:847–887. doi:10.1007/s00425-015-2452-8.
- Dawson IK, Guariguata MR, Loo J, Weber JCR, Lengkeek A, Bush D, Cornelius J, Guarino L, Kindt R, Russell J, et al. 2013. What is the relevance of smallholders’ agroforestry systems for conserving tropical tree species and genetic diversity in circa situm, in situ and ex situ settings? A review. Biodiversity and Conservation. 22:301–324.
- Doran J, Bush D, Page T, Glencross K, Sethy M, Viji I. 2012. Variation in growth traits and wood density in whitewood (Endospermum medullosum): a major timber species in Vanuatu. International Forestry Review. 14:476–485. doi:10.1505/146554812804715946.
- Doran J, Thomson L, Brophy J, Goldsack B, Bulai P, Faka’osi T, Mokoia T. 2005. Variation in heartwood oil composition of young sandalwood trees in the south Pacific (Santalum yasi, S. album and F1 hybrids in Fiji, and S. yasi in Tonga and Niue). Sandalwood Research Newsletter. 20:3–7.
- Dowell A. 2020. Essential oil of Santalum austrocaledonicum: developing an international standard. In: Page T, Meadows J, editors. Sandalwood Regional Forum; 2019 Nov 11–13; Canberra (Australia): Australian Centre for International Agricultural Research.
- Elevitch CR. 2006. Traditional trees of Pacific islands: their culture, environment and use. Holualoa (HI): Permanent Agriculture Resources.
- Ellis MF, Sedgley M. 1992. Floral morphology and breeding system of three species of Eucalyptus, section Bisectaria (Myrtaceae). Australian Journal of Botany. 40:249–262. doi:10.1071/BT9920249.
- Grattapaglia D, Silva-Junior OB, Resende RT, Cappa EP, Müller BSF, Tan B, Isik F, Ratcliffe B, El-Kassaby YA. 2018. Quantitative genetics and genomics converge to accelerate forest tree breeding. Frontiers in Plant Science. 9:1693. doi:10.3389/fpls.2018.01693.
- Griffin AR, Potts BM, Vaillancourt RE, Bell JC. 2019. Life cycle expression of inbreeding depression in Eucalyptus regnans and inter-generational stability of its mixed mating system. Annals of Botany. 124:179–187. doi:10.1093/aob/mcz059.
- Harbaugh DT. 2007. A taxonomic revision of Australian northern sandalwood (Santalum lanceolatum, Santalaceae). Australian Systematic Botany. 20:409–416. doi:10.1071/SB07009.
- Harbaugh DT, Baldwin BG. 2007. Phylogeny and biogeography of the sandalwoods (Santalum, Santalaceae): repeated dispersals throughout the Pacific. American Journal of Botany. 94:1028–1040. doi:10.3732/ajb.94.6.1028.
- Harwood C, Bulman P, Bush D, Mazanec R, Stackpole D. 2001. Australian Low Rainfall Tree Improvement Group: compendium of hardwood breeding strategies. Canberra (Australia): Rural Industries Research and Development Corporation. RIRDC Publication 01/100.
- Harwood CE, Bush DJ, Butcher T, Bird R, Henson M, Lott R, Shaw S. 2007. Achievements in forest tree genetic improvement in Australia and New Zealand. 4. Tree improvement for low-rainfall farm forestry. Australian Forestry. 70:23–27. doi:10.1080/00049158.2007.10676258.
- Huish RD, Faka’osi T, Likiafu H, Mateboto J, Huish KH. 2015. Distribution, population structure, and management of a rare sandalwood (Santalum yasi, Santalaceae) in Fiji and Tonga. Pacific Conservation Biology. 21:27–37. doi:10.1071/PC14902.
- ISO. 2002. Oil of sandalwood (Santalum album L.). Geneva: International Organization for Standardization; p. 6.
- IUCN. 2012. IUCN Red List Categories and Criteria: version 3.1. 2nd ed. Cambridge (UK): International Union for Conservation of Nature.
- Jones CG, Ghisalberti EL, Plummer JA, Barbour EL. 2006. Quantitative co-occurrence of sesquiterpenes; a tool for elucidating their biosynthesis in Indian sandalwood, Santalum album. Phytochemistry. 67:2463–2468. doi:10.1016/j.phytochem.2006.09.013.
- Jones CG, Keeling CI, Ghisalberti EL, Barbour EL, Plummer JA, Bohlmann J. 2008. Isolation of cDNAs and functional characterisation of two multi-product terpene synthase enzymes from sandalwood, Santalum album L. Archives of Biochemistry and Biophysics. 477:121–130. doi:10.1016/j.abb.2008.05.008.
- Jyothi PV, Alturi JB, Subba RC. 1991. Pollination ecology of Santalum album (Santalaceae). Tropical Ecology. 32:92–104.
- Lindgren D. 2003. Low-input tree breeding strategies. In: Wei R-P, Xu D, editors. Eucalyptus plantations: research, management and development. Singapore: World Scientific Publishing Company; p. 149–166.
- Ma G-H, Bunn E, Zhang J-F, Wu G-J. 2006. Evidence of dichogamy in Santalum album L. Journal of Integrative Plant Biology. 48:300–306. doi:10.1111/j.1744-7909.2006.00201.x.
- Mahesh HB, Subba P, Advani J, Shirke MD, Loganathan RM, Chandana S, Shilpa S, Chatterjee O, Pinto SM, Prasad K, et al. 2018. Multi-omics driven assembly and annotation of the sandalwood (Santalum album) genome. Plant Physiology. 176:2772–2788. doi:10.1104/pp.17.01764.
- Mazanec RA, Grayling PM, Spencer B, Doran J, Neumann C. 2017. Provenance variation, genetic parameters and potential gains from breeding for biomass and cineole production in three-year-old Eucalyptus loxophleba subsp. lissophloia progeny trials. Australian Forestry. 80:34–42. doi:10.1080/00049158.2016.1275100.
- McComb JA. 2009. Clonal Santalum album growth, oil content and composition on different hosts and at different locations. Journal of the Royal Society of Western Australia. 92:15–25.
- Motuliki P. 2020. Tonga sandalwood. In: Page T, editor. Sandalwood Regional Forum; 2019 Nov 11–13; Canberra (Australia): Australian Centre for International Agricultural Research.
- Muir K, Byrne M, Barbour E, Cox MC, Fox JED. 2007. High levels of outcrossing in a family trial of Western Australian sandalwood (Santalum spicatum). Silvae Genetica. 56:222. doi:10.1515/sg-2007-0033.
- Ouyang Y, Zhang XH, Chen YL, da Silva JAT, Ma GH. 2016. Growth, photosynthesis and haustorial development of semiparasitic Santalum album L. penetrating into roots of three hosts: a comparative study. Trees – Structure and Function. 30:317–328. doi:10.1007/s00468-015-1303-3.
- Page T, Bush D, Sethy M, Doran J. 2017. Variation in early growth in a second-generation whitewood (Endospermum medullosum) progeny trial in Vanuatu. Australian Forestry. 80:121–126. doi:10.1080/00049158.2017.1321462.
- Page T, Doran J, Tungon J, Tabi M. 2020. Participatory domestication strategy for Santalum austrocaledonicum in Vanuatu. Australian Forestry. 83(4):216–228.
- Page T, Hanington T, Tungon J, Tabi M, Kamasteia P. 2012. Vanuatu sandalwood growers’ guide for sandalwood production in Vanuatu. Canberra (Australia): Australian Centre for International Agricultural Research.
- Page T, Jeffrey G, Macdonell P, Hettiarachichi D, Boyce MC, Lata A, Oa L, Rome G. 2020. Morphological and heartwood variation of Santalum macgregorii in Papua New Guinea. Australian Forestry. 83(4):195–207.
- Radomiljac A, McComb J, Pate J. 1999. Gas exchange and water relations of the root hemi-parasite Santalum album L. in association with legume and non-legume hosts. Annals of Botany. 83:215–224. doi:10.1006/anbo.1998.0815.
- Robson K, Barnes M. 2020. Challenges for sandalwood plantation development in tropical Australia. In: Page T, Meadows J, editors. Sandalwood Regional Forum; 2019 Nov 11–13; Canberra (Australia): Australian Centre for International Agricultural Research.
- Rocha D, Ashokan PK, Santhoshkumar AV, Anoop EV, Sureshkumar P. 2017. Anatomy and functional status of haustoria in field grown sandalwood tree (Santalum album L.). Current Science. 113:130–133. doi:10.18520/cs/v113/i01/130-133.
- Rugkhla A, McComb JA, Jones MGK. 1997. Intra- and inter-specific pollination of Santalum spicatum and S. album. Australian Journal of Botany. 45:1083–1095. doi:10.1071/BT96079.
- Santos WD, Souza DCL, Moraes MLTD, Aguiar AVD. 2016. Genetic variation of wood and resin production in Pinus caribaea var. hondurensis Barret & Golfari. Silvae Genetica. 65:31–37. doi:10.1515/sg-2016-0004.
- Sedgley M, Griffin AR. 1989. Sexual reproduction of tree crops. Sydney (Australia): Academic Press.
- Tamla HT, Cornelius JP, Page T. 2012. Reproductive biology of three commercially valuable Santalum species: development of flowers and inflorescences, breeding systems, and interspecific crossability. Euphytica. 184:323–333. doi:10.1007/s10681-011-0530-y.
- Thomson LAJ. 2006. Santalum austrocaledonicum and S. yasi (sandalwood). In: Elevitch CR, editor. Species profiles for Pacific island agroforestry. Holualoa (HI): Permanent Agriculture Resources; p. 21.
- Thomson L. 2020. Looking ahead – global sandalwood production and markets in 2040, and implications for Pacific Island producers. Australian Forestry. 83(4):245–254.
- Thomson L, Bush D, Bulai P. 2018. Species accounts: Santalum yasi. In: Thomson L, Doran J, Clarke B, editors. Trees for life in Oceania. Canberra (Australia): Australian Centre for International Agricultural Research; p. 210–213.
- Thomson L, Bush D, Lesubula M. 2020. Participatory value chain study for yasi sandalwood (Santalum yasi) in Fiji. Australian Forestry. 83(4):227–237.
- White TL, Adams WT, Neale DB. 2007. Forest genetics. Wallingford (UK): CABI Publishing.
- Williams CG, Savolainen O. 1996. Inbreeding depression in conifers: implications for breeding strategy. Forest Science. 42:102–117.
