Abstract
Karyotype analysis of the seven varieties of Vicia faba L., namely BPL-4070, HBP-A, L-82009, IC-526385, IC-424900, IC-391651 and IC-361499 procured from Indian Agricultural Research Institute, New Delhi, was carried out to ascertain whether varietal differences are marked by chromosomal features. All the varieties exhibited diploid status with 2n = 12 chromosomes each, with perceptible differences in the conventional karyotypic features, total chromatin length (TCL) and the total form percentage (TF%). The assessment revealed the asymmetric nature of the karyotypes; telocentric chromosomes are most frequent, representing the advanced status of the V. faba varieties studied. A pair of satellite chromosome was observed in mitotic metaphase in all the varieties except L-82009, in which secondary constriction could not be discerned.
Introduction
The genus Vicia L. (Fabaceae), consisting of 166 species, is distributed mainly in Europe, Asia and North America, extending to the temperate regions of South America and tropical Africa (Maxted Citation1993). About 40 species chiefly of European origin are cultivated as vegetable crops; sometimes grown for green manure, but more often for stock feed. The feed value of broad beans is high; it is considered, in some areas, to be superior to field peas or other legumes. It is one of the most important winter crops for human consumption in the Middle East. While 94% species are diploid, of which 53% species have the basic chromosome number x = 7, 33% have x = 6 and the remaining 8% have x = 5, only a small fraction (6%) of species are known to be polyploid (all tetraploid, based either on x = 6 or x = 7) (Arslan et al. 2012). Speciation and evolution of the genus Vicia was visualized to have been accompanied by variation in chromosome size and number, and the nuclear DNA content (Chool Citation1971; Raina and Narayan Citation1984).
Karyotypic differences are reliable features for taxonomy and phylogeny. Consequently, a number of studies were undertaken in the past on various Vicia faba species and their close wild relatives, to determine karyotypic differences and to understand the process of evolutionary divergence in the genus. Karyotypes of 27 species (2n = 10, 12, 14) of the genus Vicia were subjected to linear determinant analysis and it was demonstrated that this statistical procedure, using karyotype data, can be used as a tool to identify plant species with multivariate analysis techniques (Venora et al. Citation2008). Recently Arslan et al. (2012) studied chromosome numbers, karyotypes and ideograms in 11 Vicia taxa (Vicia faba not included) naturally distributed in Turkey, and determined 2n = 10, 12, 14, 24 chromosomes. Kotseruba et al. (Citation2000), while studying several widely distributed species of the genus Vicia, determined the banding pattern of mitotic metaphase chromosomes and nuclear DNA content (2C value) in root meristem cells of a Russian accession of V. faba. Based on chromosome counts and the nuclear DNA amounts (2C) in 54 diploid and two tetraploid Vicia species while the species fell into 3 groups in respect of basic chromosome number with x=5, 6 or 7, the DNA amount ranged from approximately 4 – 27 picograms, the highest value represented by Vicia faba. It was further noticed that DNA amount was not correlated with chromosome number, and there was no connection between DNA amounts and taxonomic groupings (Seal and Rees 1982).
Crop improvement programmes involve combining the beneficial characters from different germplasms into a single form that suits the agroclimatic conditions of a particular region, so that the quality of the produce is enhanced. Karyotype analysis is a prerequisite for designing the breeding programmes. Since the majority of present-day cultivars are patented, it is important to be able to identify them. Knowledge of the karyotypic features of species, i.e. the number of chromosomes, relative size differences among chromosomes of a complement and the morphology and behaviour of the chromosomes during mitosis and meiosis, is necessary for comparative assessment of the genomic compatibility between species/varieties, so that the crop plant can be improved by conventional breeding. Hybridization at inter-varietal level, where there is closer genomic similarity, is mostly attempted to enhance the yield of crops. Desirable hereditary traits, such as resistance to disease, drought, insects and frost, that are commonly found in the wild relatives of cultivated plants are screened for and conserved for use in hybridization programmes (Tabete Citation1927; Omidiji Citation1975).
The present karyotype analysis of seven varieties of V. faba L. (BPL-4070, HBP-A, L-82009, IC-526385, IC-424900, IC-391651 and IC-361499) was performed to understand the taxonomic status and evolutionary trends in the genus, to enhance the scope for gene manipulation, and thus to qualitatively and quantitatively improve the crop. This type of varietal demarcation on the basis of karyotype analysis has previously been performed in other plants (Koo et al. Citation2002; Ines and Farnándes 2004; Parvin et al. Citation2008).
Material and methods
Squash preparations of meristem root tip cells of V. faba varieties procured from Indian Agricultural Research Institute, New Delhi were made for mitotic studies, following the procedures given by Sharma and Sharma (Citation1990), using 2% acetocarmine as nuclear stain. For karyotype analysis, well spread metaphase squashes were selected from temporary preparations and chromosome measurements were taken with the help of calibrated ocular micrometer and/or camera lucida drawings of chromosomes made at bench level from temporary slides. For the purpose of the ideogram the haploid chromosomes were designated as 1–6 and arranged in decreasing order of size. To provide greater resolution of the chromosome structural alterations during varietal divergence, chromosomes were classified following Abraham and Prasad (Citation1983) using the following terms: nearly median, nearly submedian (–), nearly submedian (+), nearly subterminal (–), and nearly subterminal (+) for chromosomes with long arm/short arm ratios of 1.01–1.63, 1.64–2.99, 3.01–4.26, 4.27–6.99 and 7.01–14.38, respectively. Relative length, arm ratio (long arm/short arm) of the chromosomes and S% (shortest chromosome/longest chromosome × 100) were determined. Total form percentage (TF%) was also calculated using the formula given by Huziwara (Citation1962).
Results
The analysed parameters and the karyotype formulae for the various V. faba varieties calculated in this study are summarized in . Microphotographs of the metaphase chromosomes, and diagrammatic representation (ideograms), are provided in , respectively.
Table 1. Chromosomes characteristics of seven varieties of Vicia faba L.
Table 2. Karyotypic details of seven varieties of Vicia faba L.
Figure 1 Microphotographs of somatic metaphase chromosomes (2n = 12) of the seven varieties of Vicia faba L. (A) BPL-4070; (B) HBP-A; (C) L-82009; (D) IC-526385; (E) IC-424900; (F) IC-391651; (G) IC-361499. Scale bars = 10 μm.
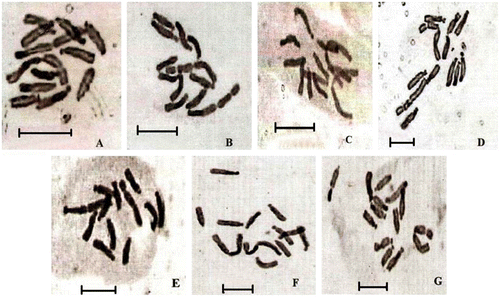
Figure 2 Ideograms of somatic metaphase chromosomes of Vicia faba L. Note the absence of a satellite arm in var. L-82009. Scale bar = 10 μm.
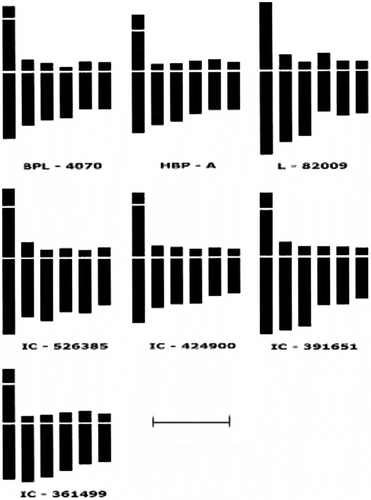
All the seven varieties of Vicia faba L. investigated in this study exhibited 2n = 12 large chromosomes, ranging from 5.66 μm in IC-424900 to 20.00 μm in L-82009. The total genomic metaphase chromosome length marginally varied in different varieties in the range of 49.81 μm to 62.00 μm. The metacentric chromosome pair was found to be longest in the complement, and consisted of a satellite on its short arm, except for variety L-82009 in which nucleolar-organizing chromosomes could not be deciphered. A perusal of chromosome data also clearly indicates the asymmetric nature (3B) (Stebbins Citation1971) of karotypes in all the varieties under study. From Table it is apparent that except for the longest nucleolar organizing (NO) chromosome pair, which is metacentric, all other pairs of chromosomes are either submetacentric or subtelocentric. The karyotype formulae (KF) of the seven varieties of V. faba summarize the gross morphological differences in the chromosomes within and between the varieties investigated.
Discussion
The grouping of varieties on the basis of karyotype characteristics and determination of the % chromosome variability may provide additional information for understanding the karyotypic relationship among different varieties. This information may be helpful in enhancing the breeding prospects of the crop. In the present investigation, cytological karyotyping of all the seven varieties of V. faba represented a common general pattern, indicating that they constitute a homogenous assemblage diverging from a single basic set of chromosomes (x = 6) (Raina and Rees Citation1983). Critical examination of these varieties revealed distinct karyotypic differences, each variety differing from the others in chromosome type, TCL (total chromatin length) and TF% (total form percentage). Such differences among varieties are in accordance with the findings of other studies, e.g. Munian and Subramanian (Citation1985), Das and Mallick (Citation1989) and Jacobkutty and Bhavanandan (Citation1996). Evaluation of the karyotypic features of different varieties of V. faba reflected no major intraspecific variation. The total chromatin length as represented by metaphase chromosomes is highest (62.00 μm) in L-82009 and lowest (49.81 μm) in HBP-A. Decrease in TCL is one of the factors indicating the trend of evolution (Stebbins 1950). Thus, HBP-A (TCL 49.81 μm) may be considered as most advanced and L-82009 (TCL 62.00 μm) as the most primitive among the varieties of V. faba studied. The difference in TCL may be attributed to chromosome aberrations, primarily deletion of inert chromatin segments (the heterochromatin parts), which seems to have occurred in nature, resulting in diminution of the chromosome sizes. It can thus be concluded that the reduction in TCL in HBP-A might be due to deletion of heterochromatin from the progenitor complement, and varietal demarcation is a result of change in heterochromatic parts and/or repetitive sequences in the functional segments of genome during evolution. It has earlier been suggested that the evolution of species and genera is accompanied by changes in the types of repetitive DNA present (Flavell et al. Citation1977) and also that amplification of DNA has a significant role in the microevolution of the genome.
The subtelocentric chromosomes in the chromosome complements of V. faba varieties suggest that the karyotype of all the seven varieties are asymmetric in nature, indicating that advanced characteristics have evolved. This finding is in agreement with those reported earlier in the species of Vicia, where moderate asymmetry and moderate symmetry have been observed (Ruijun and Haoyou Citation1994, Citation1995). Based on TF%, IC-391651 exhibits the most asymmetrical chromosomes, having the lowest TF% value (23.72%), followed by IC-526385 (24.70%), IC-361499 (25.00%), HBP-A (25.31%), L-82009 and BPL-4070 (26.88%), and IC-424900 (27.55%). From this point of view IC-391651 may be considered as the most advanced, and IC-424900 as the most primitive variety. The reduction in the size of some chromosomes in progressively unequal sizes may account for a shift in asymmetrical karyotypes from symmetrical ones during progressive evolution, and the reduction in length of one arm of the chromosomes may be responsible for derivation of submedian and subterminal centromeric chromosomes from median ones. The other significant variation in the karyotype is shift in the position of centromere from median to submedian to subterminal in the reconstructed karyotypes, owing to chromosomal rearrangements. Accumulation of differences in the relative sizes between chromosomes of the same complement and gradual evolution from symmetrical to asymmetrical karyotype have been elegantly discussed by Stebbins (Citation1971) and Sharma (Citation1976). Though the exact mechanism of origin of these karyotypic differences is difficult to determine, the changes may have helped the process of evolution and the emergence of different varieties. On the other hand, the uniformity in the chromosome number and considerable similarity in gross characteristics of the cytological karyotypes suggest gene mutations to be the principal factor in evolution. In this context, Goswami’s (Citation1979) report about recent origin of the varieties of black gram (Phaseolus mungo) by studying the karyotypes of 32 varieties is noteworthy. The absence of satellites in a pair of chromosomes in L-82009 may be due either to breakage of the satellite arm and its subsequent loss, or to deletion of the secondary constriction region followed by reunion of the broken ends of the chromosome during evolution.
The ratio of longest to shortest chromosome lengths indicates chromosome length variation in relation to relative phylogenetic status among the varieties. The highest value (3.07) is recorded in L-82009 and the lowest (2.28) in IC-361499 (Table ). Based on TCL, L-82009, IC-526385 and IC-391651 exhibit greater resemblance to each other and thus more advanced evolutionary status than the other four varieties, HBP-A, IC-424900, IC-361499 and BPL-4070, which have lower TCL values. The overall assessment of the chromosome characteristics does not reveal marked karyotypic variation, yet they seem sufficient to cause varietal differentiation in V. faba L.
Acknowledgements
The authors would like to thank the authorities of IARI, New Delhi for courtesy in providing the seed germplasms of V. faba varieties. Financial assistance provided in the form of UGC-BHU fellowship is also gratefully acknowledged.
References
- Abraham , Z and Prasad , N . 1983 . A system of chromosome classification and nomenclature . Cytologia , 48 ( 1 ) : 95 – 101 .
- Arslan , E , Ertugrul , K and Ozturk , AB . 2012 . Karyological studies of some species of the genus Vicia L. (Leguminosae) in Turkey . Caryologia , 65 ( 2 ) : 106 – 113 .
- Chool , WY . 1971 . Variation in nuclear DNA content in the genus Vicia . Genetics , 68 ( 2 ) : 195 – 211 .
- Das , AB and Mallick , R . 1989 . Variation in karyotype and nuclear DNA content in different varieties in Foeniculum vulgare Mill . Cytologia , 54 ( 1 ) : 129 – 134 .
- Flavell , RB , Rimpau , J and Smith , DB . 1977 . Repeated sequence DNA relationships in four cereal genomes . Chromosoma , 63 ( 3 ) : 205 – 222 .
- Goswami , LC . 1979 . Karyological studies of 32 varieties of black gram Phaseolus mungo L . Cytologia , 44 ( 3 ) : 549 – 556 .
- Huziwara , Y . 1962 . Karyotype analysis in some genera of Aster . Am J Bot , 49 ( 2 ) : 116 – 119 .
- Ines , LG and Farnándes , A . 2004 . Karyotype studies in Arachis hypogeae L. varieties . Caryologia , 57 ( 4 ) : 353 – 359 .
- Jacobkutty , MJ and Bhavanandan , KV . 1996 . Karyomorphological studies in Allium sativum . J Cytol Genet , 31 ( 3 ) : 49 – 54 .
- Koo , DH , Hurr , YK , Jin , DC and Bang , JW . 2002 . Karyotype analysis of a Korean cucumber cultivar (Cucumis sativas L. cv. Winter Long) using C-banding and bicolor fluorescence in situ hybridization . Mol Cell , 13 ( 3 ) : 413 – 418 .
- Kotseruba , VV , Venora , G , Blangiforti , S , Castiglione , MR and Cremonini , R . 2000 . Cytology of Vicia species. IX. Nuclear DNA amount, chromatin organization and computer aided karyotyping of a Russian accession of Vicia faba L . Caryologia , 53 ( 3–4 ) : 195 – 204 .
- Maxted , N. 1993 . A phenetic investigation of Vicia (Leguminosiae, Vicieae) . Bot J Linn Society , 111 : 155 – 182 .
- Munian , M and Subramanian , D . 1985 . Karyological studies of sixteen varieties of Tepary bean Phaseolus acutifolius A . Gray. Cytologia , 50 : 31 – 37 .
- Omidiji , MO . 1975 . Interspecific hybridization in the cultivated non-tuberous Solanum species . Euphytica , 24 : 341 – 353 .
- Parvin , S , Kabir , G , Ud-Deen , MM and Sarker , JK . 2008 . Karyotype analysis of seven varieties of Taro Colocasia esculenta (L.) Schott. from Bangladesh . J Bio Sci , 16 : 15 – 18 .
- Raina , SN and Narayan , RKJ . 1984 . Changes in DNA composition in the evolution of Vicia species . Theor Appl Genet , 68 ( 1–2 ) : 187 – 192 .
- Raina , SN and Rees , H . 1983 . DNA variation between and within chromosome complements of Vicia spp . Heredity , 51 ( 1 ) : 335 – 346 .
- Ruijun , L and Haoyou , W . 1994 . Karyotype analysis in Vicia venosa Willd. Ex. Link Maxim. and V. ramuliflora Maxim . Ohwi. Cytologia , 59 ( 3 ) : 361 – 364 .
- Ruijun , L and Haoyou , W . 1995 . Karyotype of Vicia sect . Vicilla from Northeast China. Cytologia , 60 ( 4 ) : 353 – 358 .
- Seal , A and Rees , H . 1982 . The distribution of quantitative DNA changes associated with the evolution of diploid Festuceae . Heredity , 49 ( 2 ) : 179 – 190 .
- Sharma , A . 1976 . The chromosomes , New Delhi : Oxford and IBH .
- Sharma , AK and Sharma , A . 1990 . Chromosomes techniques: Theory and practice , 3rd edition , London : Butterworths .
- Stebbins GL.1950. Variation and evolution in plants. New York: Columbia University Press.
- Stebbins , GL. 1971 . Chromosomal evolution in higher plants , London : Edward Arnold .
- Tabete , T . 1927 . On the first generation hybrid of egg plant . CbI. Hort Japan , 187 : 19 – 21 .
- Venora , G , Ravalli , C and Cremonini , R . 2008 . The karyotype as a tool to identify plant species: Vicia species belonging to Vicia subgenus . Caryologia , 61 ( 3 ) : 300 – 319 .