Abstract
Avian karyotypes are remarkably conserved throughout evolution. Hence, studies have given priority to the chicken for characterizing and understanding the chromosome organization in this clade. Chicken chromosomes are conventionally classified as macrochromosomes (MACs), microchromosomes (MICs) and sexual chromosomes (ZW). For expanding karyotype data, the DNA amount of the MACs, ZW and a few MICs have been reported. In spite of the progress, hitherto chicken chromosomes have not been fully classified and characterized, especially MICs, accounting for a gap in evolutionary approaches and genomic projects. The chicken karyotype is considered one of the most challenging for cytogenetic study. This study focused on image cytometry, a quantitative measure of the chromosome DNA amount, to fine-tune morphometric data. The methodology was fundamental to identify and discriminate each MIC, overlapping between MICs and MICs/MACs. Associating these data to classical cytogenetic rules, karyograms were assembled. Mean DNA amount was also used to calculate the percentage equivalent of each chromosome group in the haploid genome. Therefore, image cytometry represented a powerful application that can be used for avian karyotype characterization, opening up a range of possibilities for scientific research.
1. Introduction
In addition to its economic importance, Gallus domesticus (domestic chicken) is a good model system for basic studies in biology (Schmid et al. Citation2005) and the focus of a genome mapping project in avians (International Chicken Genome Sequencing Consortium Citation2004). Therefore, the karyotype of this species has been analyzed in the last three decades (Stubblefield and Oro Citation1982; Auer et al., Citation1987, Tiersch and Wachtel Citation1991; Fillon et al. Citation1998; Ladjali-Mohammedi et al. Citation1999; Andreozzi et al. Citation2001; Grutzner et al. Citation2001; Masabanda et al., Citation2004; Ellegren Citation2005).
The chromosome number of chicken was estimated as 2n = 78 by Pollock and Fechheimer (Citation1976), showing the sex chromosomes Z and W and female as a heterogametic sex (Auer et al. Citation1987; Nakamura et al. Citation1990; Bloom et al. Citation1993; Smith and Burt Citation1998; Ladjali-Mohammedi et al., Citation1999; Masabanda et al., Citation2004). The chicken chromosome has also been analyzed by banding (as revised by Auer et al., Citation1987; Schimid et al. Citation1989; Ladjali-Mohammedi et al., Citation1999) and molecular cytogenetic techniques (Andreozzi et al. Citation2001; Masabanda et al. Citation2004). Based on these data, the chicken karyotype has been conventionally classified as showing macrochromosomes (MACs), microchromosomes (MICs), and sexual Z and W chromosomes.
Owing to the continued progress of cytogenetic approaches, some authors have proposed models for classification of chicken chromosomes, which have been extrapolated for other avian species. Using the total length as parameter, Bloom et al. (Citation1993) suggested the first classification for chicken chromosomes: 1–4 and Z were considered large chromosomes, and thus denoted MACs; 5–9 and W as intermediate or large MICs; and 10–38 as MICs. The last classification was shown by Masabanda et al. (Citation2004) through molecular cytogenetic and flow-karyotyping data. According to these authors, chromosomes 1–10 plus ZW belong to the group A, 11–16 (with 16 exhibiting the NOR) to the group B, 17–32 to the group C, and 33–38 to the group D.
In addition to qualitative approaches, quantitative analyses were also performed on the chicken karyotype. Image cytometry (ICM) was used for nuclear DNA amount measurement in chicken, agreeing with flow cytometry data (Tiersch and Wachtel Citation1991). The female chicken nuclear genome showed 2C = 2.15 picograms (pg) of DNA, equivalent to 1C = 1.05 × 109 base pairs (bp), while the male had 2C = 2.24 pg, 1C = 1.095 × 109 bp (Mendonça et al. Citation2010).
As well as the nuclear DNA content, the chromosomal DNA amount was measured. Bloom et al. (Citation1993) used the chromosome area parameter and calculated the DNA amount of the chromosomes 1–9, 16, 19, 29, Z and W. Smith and Burt (Citation1998) used the fluorescence intensity of propidium iodide to measure the DNA amount of the chromosomes 1–8, Z and W. The DNA amount of the chicken chromosomes (1–28, Z and W) was also measured in genome sequencing project (EMSEMBL, release 83; Kersey et al. Citation2015; Table ).
Table 1. 1C DNA amount of chicken chromosomes.
In spite of all these advances, so far chicken chromosomes have not been fully classified and characterized, especially MICs, which are considered extremely difficult to analyze (Kaelbling and Fechheimer Citation1983; Fillon et al., Citation1998; Grutzner et al., Citation2001; Bloom et al Citation1993; Ladjali-Mohammedi et al., Citation1999; Masabanda et al., Citation2004). This fact has hindered understanding the genome organization in avians, representing a significant barrier to the progress of genome mapping and evolution studies (Masabanda et al., Citation2004; Schmid et al., Citation2005), making the chicken karyotype one of the most challenging for cytogenetic study.
As ICM has resolved the DNA amount in species showing relatively small chromosomes (Carvalho et al. Citation2011), the present study was based on one question: would ICM be an appropriate tool for measuring the DNA amount of all chicken chromosomes?
2. Material and methods
2.1. Biological material
Chickens (Gallus gallus domesticus ‘Leghorn’) were kindly supplied by Dr Paulo César Gomes (Animal Science Department, Universidade Federal de Viçosa (UFV), Brazil). All experimental procedures were conducted under the supervision of the Animal Care and Use Committee of the UFV.
2.2. Chromosome slide preparation
Metaphase cells were obtained from bone marrow of female and male chickens after colchicine treatment (Macgregor and Varley Citation1988). Bone marrow cells were collected from both femora by flushing into Ringer’s solution, and cells were centrifuged at 100 × g for 5 min. The supernatant was discarded and the cells were hypotonized with 0.075 M KCl, incubated at 37 °C for 10 min, and centrifuged at 100 × g for 5 min. The supernatant was poured out, and the pellet was fixed with cold methanol-acetic acid (Merck, Darmstadt, Germany) 3:1 (v/v). Fixation was repeated twice, with an interval of 10 min, and the suspensions were stored at –20 °C. The cells were resuspended in fresh fixative solution, and two to three drops were dripped onto a clean slide and placed on a 50 °C hot plate to dry.
2.3. Feulgen reaction
All slides were stored in 70% ethanol (Merck) at –20 °C for 24 h. The Feulgen reaction was performed according to Carvalho et al. (Citation2011). The slides were post-fixed in 4% formaldehyde solution (Merck) for 60 min, washed in distilled water, air dried, and hydrolyzed in 5 M HCl (Merck) for 10–60 min at 25 °C. After the hydrolysis step, slides were stained with Schiff’s reagent (Merck) for 12–24 h at 4 °C. Finally, slides were washed for three times of 3 min in 0.5% SO2-water (Merck) and in distilled water.
2.4. Microscope and image analysis system instrumentation
Metaphase images were captured with a monochromatic charge-coupled device digital video camera of 12-bit gray and a frame-grabber card (Photometrics CoolSNAP Pro-Roper Scientific; Tucson, AZ, USA). This camera was assembled on a trinocular photomicroscope (Olympus BX-60; Center Valley, PA, USA) equipped with: a source of stabilized light; UPlanFI objective with 40 × magnification and 0.75 numeric aperture; PlanApo 100 × oil immersion objective with 1.40 numeric aperture; aplanat achromat condenser with 1.4 aperture; and neutral density filter (ND6). The video camera was further coupled to a Pentium 4 HT computer (Dell Optiplex GX 620; Round Rock, TX, USA) containing Image Pro-Plus 6.1 software (Media Cybernetics; Bethesda, MD, USA). Calibration and setup of the image analysis system were accomplished following the procedure described by Mendonça et al. (Citation2010) and Carvalho et al. (Citation2011) for ICM measurement.
The 12-bit gray images of each metaphase were captured using the Image Pro-Plus 6.1 software tools. The images were selected, segmented and cut, using the digital tools to assemble the karyotypes.
2.5. Chromosomal DNA amount and statistical analyses
The mean DNA amount of each chicken chromosome, in pg, was measured by proportionally distributing the nuclear 4C-value, according to the data obtained by Mendonça et al. (Citation2010) using nuclei of chicken red blood cells (CRBC). The values were calculated according to Carvalho et al. (Citation2011). The mean values were converted into bp, considering that 1 pg of DNA corresponds to 0.978 × 109 bp (Doležel et al. Citation2003).
Mean values for chromosomal DNA amount of females were correlated with those of males by Pearson linear correlation and regression analysis. These statistical analyses were carried out using the statistical software Genes (Cruz Citation2013).
3. Results
Cytogenetic preparations, hydrolyzed with 5 M HCl solution for 45 min and incubated in Schiff’s reagent for 12 h, presented stoichiometrically stained chromosomes. These slides also exhibited metaphases showing other fundamental aspects for ICM execution: chromosomes well-spread and morphologically preserved, flattened on the slide, without cytoplasmic background and structural deformations of chromatin. From these metaphases, 10 were chosen for male and 10 for female. All karyotypes showed 76 autosomal and two (ZZ or ZW) sexual chromosomes.
The precise delineation of all chromosomes was performed using image analysis programs. After morphometric characterization (total and arm lengths), the integrated optical density (IOD) value was automatically calculated for each chromosome of the male and female karyotypes. From IOD values and nuclear genome size (2C = 2.15 pg for female or 2C = 2.24 pg for male), the mean DNA amount of each chromosome could be measured.
DNA amount values of each chromosome were statistically compared, showing that mean values of the 38 autosomal and Z chromosomes of male fully correlated with those of female (R2 = 99.99%, Figure ). Considering this result, the DNA amount value of each chromosome from male was added to those of female, totaling 40 values for each autosomal chromosome, 30 for Z, and 10 for W. So, the mean DNA amount values were measured (Table ), ranging from 1C = 0.158 (chromosome 1) to 0.003 pg (38), 0.041 (Z), and 0.014 pg (W). Associating classical cytogenetic rules, morphometric and ICM data, 10 karyograms were assembled for male and 10 for female (Figure ).
Figure 1. Relationship between the DNA mean values of each male (axis x) and female (axis y) chicken chromosomes measured from ICM. For Pearson linear correlation and regression analysis, the mean values of DNA amount were multiplied by 1000.
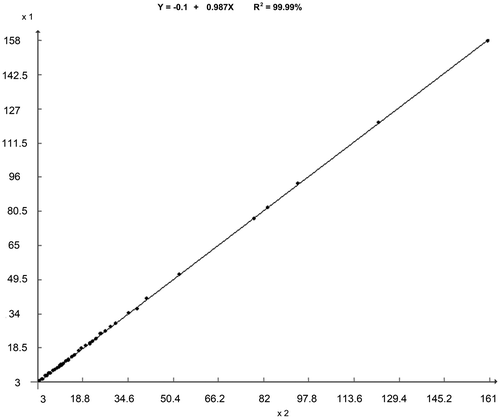
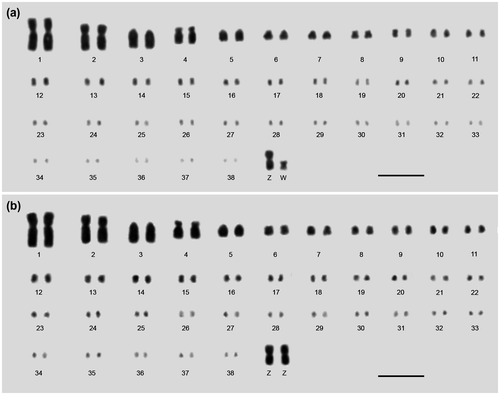
Figure 2. Representative karyogram of (a) the female and (b) the male chicken assembled from metaphasic chromosomes stained from Feulgen reaction. The criteria established in the 4th European Colloquium of Cytogenetics of Domestic Animals and the chromosomal DNA amount were considered to consecutively order and number the chromosomes. The karyograms showed 76 autosomal and ZZ (male) or ZW (female) sexual chromosomes. Bar = 5 μm.
4. Discussion
Chicken metaphases showed chromosomes with sufficient resolution for precise delineation by digital segmentation, which is a fundamental step for cytogenetic characterization and ICM accomplishment. The mean chromosomal DNA amount was separately measured for each chromosome of female and male. Statistical analysis revealed high correlation (R2 = 99.99%) between the mean values of each of the 38 autosomal and Z chromosomes. This result showed that the cytogenetic and ICM procedures, as well as setup and calibration of the image analysis system, were accurate.
From morphometric and chromosome DNA amount data, 20 karyograms were assembled, 10 for male and 10 for female. All chicken karyograms showed 2n = 78, including MACs, several MICs, and sexual chromosomes, with ZZ for male and ZW for female. The karyograms were assembled agreeing to criteria established in the 4th European Colloquium of Cytogenetics of Domestic Animals (Ladjali-Mohammedi et al., Citation1999). Besides total length, the chromosomal DNA amount was considered to consecutively order and number the chromosomes.
The adopted procedures and tools were fundamental to identify each MIC, overlapping between MICs and MICs/MACs. In spite of the relative difficulty of establishing centromere and telomere regions of the MICs, the total length and DNA amount were measured and these chromosomes were paired. As for several birds, the very small size and large number of MICs have often hampered chicken karyotype characterization (Kaelbling and Fechheimer Citation1983; Fillon et al., Citation1998; Bloom et al., Citation1993; Ladjali-Mohammedi et al., Citation1999; Grutzner et al., Citation2001; Masabanda et al., Citation2004), only allowing access to the first 15 autosomal and ZW chromosomes for more detailed study (Bloom et al., Citation1993; Ladjali-Mohammedi et al., Citation1999; Masabanda et al., Citation2004). As in these cytogenetic studies, the chicken genome assembled for some MICs (Table ) is not complete, even in the latest build (ICGC Gallus_gallus_4.0, November 2011; aka galGal4.0; Miller et al., Citation2014). These approaches have been hampered due to structural characteristics of the MICs: high GC amount and recombination rates, many tandem repeats and large numbers of gene family members (Miller et al. Citation2014).
In order to overcome the barriers for identification and mapping of the MICs, as in this study, Gaginskaya et al. (Citation2009) accomplished high resolution cytogenetic analysis using avian lampbrush chromosomes. These authors constructed a map which reflected the chromomere distribution, the average loop length and positions of landmarks. So, it was possible to individualize each chromosome and recognize the putative genes located in each MIC.
Besides of the recognition and characterization, ICM analyses of chicken karyotype showed that some chromosomes possess identical chromosomal DNA amount (Table ), such as: 14 and 15 (0.021 pg), 19 and 20 (0.015 pg), 21 and 22 (0.013 pg), 23–26 (0.011 pg), 27 and 28 (0.010 pg), 31 and 32 (0.007 pg), 33 and 34 (0.006 pg), and 35–37 (0.004 pg). Therefore, the largest differences between the values of DNA amount were found among the MACs.
As summarized in Table , distinct values of DNA amount were reported for some chicken chromosomes, especially MACs. However, other methods have been applied to measure the DNA amount, such as the chromosome area (Bloom et al., Citation1993) and propidium iodide fluorescence intensity (Smith and Burt Citation1998). Unlike these methods, ICM is a densitometric technique useful for quantitative applications (Carvalho et al., Citation2011) in several organisms, including birds (Mendonça et al., Citation2010). ICM is conducted following medical quality parameters for accurate calibration and setup of the microscopy, digital CCD camera and image analysis software, and for obtaining nuclei or chromosomes stoichiometrically stained during the Feulgen reaction. Due to some aspects of ICM, it was possible to measure the DNA amount of all chicken chromosomes, including all MICs.
Considering the chicken chromosome classification reported by Masabanda et al. (Citation2004) and the ICM results, group A (chromosomes 1–10 and ZW, the MACs) represents 67.50% of the female haploid genome, group B (11–16, the large MICs) 12.66%, group C (17–32, the small MICs) 17.32%, and group D (chromosomes 33–38, the smallest MICs) 2.52%. According to Bloom et al. (Citation1993), chromosomes 1–8 plus ZW (MACs) corresponded to 72% of the female haploid genome. For the same chromosomes, Smith and Burt (Citation1998) found 82%, and in this study the value was 62.05%. According to chicken genome sequencing project (Kersey et al., Citation2015), group A represents 82.91%, group B 8.47%, and group C (chromosomes 17–28) equivalent to 8.61%. Therefore, the values reported by Bloom et al. (Citation1993), Smith and Burt (Citation1998) and Kersey et al. (Citation2015) are overestimated in relation to the ICM data, reflecting the differences among the methods. Still, ICM evidenced that the chicken DNA amount is constituted by MICs. Therefore, the measurement of the DNA amount of the MICs, mainly chromosomes 29–38, is fundamental to tracking the real contribution of individual and chromosome groups in the genome.
For the first time, the DNA amount was reported for all chicken chromosomes. This data is considered useful for genetic mapping and evolutionary approaches. Because chicken is a good model and its karyotype is similar to other avians, the chromosome DNA data also represents a base for approaches in other species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding information
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, DF, Brazil), Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG, Belo Horizonte, MG, Brazil), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, DF, Brazil).
References
- Andreozzi L, Federico C, Motta S, Saccone S, Sazanova AL, Sazanov AA, Smirnov AF, Galkina SA, Lukina NA, Rodionov AV, et al. 2001. Compositional mapping of chicken chromosomes and identification of the gene-richest regions. Chromosome Res. 9(7):521–532. doi:10.1023/A:1012436900788.
- Auer H, Mayr B, Lambrou M, Schleger W. 1987. An extended chicken karyotype, including the NOR chromosome. Cytogenet Cell Genet. 45(3–4):218–221. doi:10.1159/000132457.
- Bloom SE, Delany ME, Muscarella DE. 1993. Constant and variable features of avian chromosomes. In: Etches RJ, Gibbon AMV, editors. Manipulation of the avian genome. 1st ed. Boca Raton: CRC Press; p. 39–59.
- Carvalho CR, Clarindo WR, Abreu IS. 2011. Image cytometry: nuclear and chromosomal DNA quantification. In: Chiarini-Garcia H, Melo RCN, editors. Light microscopy: methods in molecular biology. Vol. 689. New York, NY: Humana Press. p. 51–68. doi: 10.1007/978-1-60761-950-5_4
- Cruz CD. 2013. Genes: a software package for analysis in experimental statistics and quantitative genetics. Acta Sci Agron. 35(3):271–276. doi:10.4025/actasciagron.v35i3.21251.
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. 2003. Nuclear DNA and genome size of trout and human. Cytometry. 51(2):127–128. doi:10.1003/cyto.a.10013.
- Ellegren H. 2005. The avian genome uncovered. Trends Ecol Evol. 20(4):180–186. doi:10.1016/j.tree.2005.01.015.
- Fillon V, Morisson M, Zoorob R, Auffray C, Douaire M, Gellin J, Vignal A. 1998. Identification of 16 chicken microchromosomes by molecular markers using two-colour fluorescence in situ hybridization (FISH). Chromosome Res. 6(4):307–313. doi:10.1002/cyto.a.10013.
- Gaginskaya E, Kulikova T, Krasikova A. 2009. Avian lampbrush chromosomes: a powerful tool for exploration of genome expression. Cytogenet Genome Res. 124(3–4):251–267. doi:10.1159/000218130.
- Grutzner F, Zend-Ajush E, Stout K, Munshe S, Niveleau A, Nanda I, Schimid M, Haaf T. 2001. Chicken microchromosomes are hypermethylated and can be identified by specific painting probes. Cytogenet Cell Genet. 93(3–4):265–269. doi:10.1159/000056996.
- International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 432:695–716. doi:10.1038/nature03154.
- Kaelbling M, Fechheimer NS. 1983. Synaptonemal complexes and the chromosome complement of the domestic fowl – Gallus domesticus. Cytogenet Cell Genet. 36(3):567–572. doi:10.1159/000131847.
- Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, et al. 2015. Ensembl Genomes 2016: more genomes, more complexity (Release 83). Nucleic Acids Res. 44(D1):D574–D580. doi:10.1093/nar/gkv1209.
- Ladjali-Mohammedi K, Bitgood JJ, Tixier-Boichard M, Ponce de Leon FA. 1999. International system for standardized avian karyotypes (ISSAK): standardized banded karyotypes of the domestic fowl (Gallus domesticus). Cytogenet Cell Genet. 86(3–4):271–276. doi:10.1159/000015318.
- Macgregor HC, Varley JM. 1998. Working with animal chromosomes. 2nd ed. Chichester and New York: John Wiley & Sons.
- Masabanda JS, Burt DW, O’Brien PCM, Vignal A, Fillon V, Walsh PS, Cox H, Tempest HG, Smith J, Habermann F, et al. 2004. Molecular cytogenetic definition of the chicken genome: the first complete avian karyotype. Genetics. 166(3):1367–1373. doi:10.1534/genetics.166.3.1367.
- Mendonça MAC, Carvalho CR, Clarindo WR. 2010. DNA content differences between male and female chicken (Gallus gallus domesticus) nuclei and Z and W chromosomes resolved by image cytometry. J Histochem Cytochem. 58(3):229–235. doi:10.1369/jhc.2009.954727.
- Miller MM, Robinson CM, Abernathy J, Goto RM, Hamilton MK, Zhou H, Delany ME. 2014. Mapping genes to chicken microchromosome 16 and discovery of olfactory and scavenger receptor genes near the major histocompatibility complex. J Hered. 105(2):203–215. doi:10.1093/jhered/est091.
- Nakamura D, Tiersch TR, Douglass M, Chandler RW. 1990. Rapid identification of sex in birds by flow cytometry. Cytogenet Cell Genet. 53(4):201–205.
- Pollock DL, Fechheimer NS. 1976. The chromosome number of Gallus domesticus. Br Poult Sci. 17(1):39–42. doi:10.1080/00071667608416247.
- Schimid M, Enderle E, Schindler D, Schempp W. 1989. Chromosome banding and DNA replication patterns in bird karyotypes. Cytogenet Cell Genet. 52:139–146. doi:10.1159/000132864.
- Schmid M, Nanda I, Burt DW. 2005. Second report on chicken genes and chromosomes. Cytogenet Genome Res. 109(3–4):415–479. doi:10.1159/000084205.
- Smith J, Burt DW. 1998. Parameters of the chicken genome (Gallus gallus). Anim Genet. 29(4):290–294. doi:10.1046/j.1365-2052.1998.00334.x.
- Stubblefield E, Oro J. 1982. The isolation of specific chicken macrochromosomes by zonal centrifugation and flow sorting. Cytometry. 2(5):273–281. doi:10.1002/cyto.990020502.
- Tiersch TR, Wachtel SS. 1991. On the evolution of genome size in birds. Heredity. 82(5):363–368. doi:10.1093/jhered/.
