Abstract
A survey on chromosome counts of different sections belonging to the genus Trigonella L. (Fabaceae) throughout the world is presented and the relationships between chromosome data of its sections and their biogeography are also discussed here. In addition, chromosome numbers of 58 populations belonging to 19 species growing in Iran are reported. All studied species except T. monantha and T. orthoceras were diploid with chromosome number 2n = 2x = 16. T. monantha with four subspecies (2n = 4x = 28, 2n = 6x = 42) and T. orthoceras (2n = 6x = 48) were polyploid taxa. Based on the results, x = 7, 8 was determined for Trigonella genus. According to the chromosome and geographical data, Iran can be regarded as the center of origin of Trigonella genus.
Keywords:
Introduction
The genus Trigonella L. is a member of the Fabaceae family. Fabaceae is the second largest family of flowering plants in the world with 650 genus and 18,000 species (Rakhee et al. Citation2004). Trigonella (Fabaceae) includes about 135 species worldwide, most distributed in the dry regions around the Mediterranean, West Asia, Europe, North and South Africa, North America, and with only two species being present in South Australia (Mabberley Citation1997).
Trigonella consists of perennial or annual herbs with pinnately trifoliate leaves, often exhaling an odor and are, like other grain legumes, important for food and medicine (Chopra et al. Citation1956; Girardon et al. Citation1989; Balodi and Rao Citation1991; Bhatti et al. Citation1996; Dangi et al. Citation2004).
In Iran there are 48 taxa of the genus included in 12 sections localized in different phytogeographical regions; some 15 (32%) taxa are endemic to Iran (Rechinger Citation1984; Hamzeh’ee Citation2000; Janighorban Citation2004; Badrzadeh and Ghafarzadeh-Namazi Citation2009; Ranjbar, Karamian and Hajmoradi Citation2012; Ranjbar, Karamian, Hajmoradi, et al. Citation2012; Ranjbar and Hajmoradi Citation2012, Citation2015, Citation2016); 14 of those are perennial species of Trigonella sect. Ellipticae (Boiss.) Sirj. Cytogenetic investigations were conducted on Trigonella by Singh and Roy (Citation1970), Singh and Singh (Citation1976), Agarwal and Gupta (Citation1983), Ahmad et al. (Citation1999), Dundas et al. (Citation2006), Martin et al. (Citation2008, Citation2010, Citation2011a, Citation2011b), Yilmaz et al. (Citation2009); Ranjbar, Hajmoradi and Karamian (Citation2011, Citation2014), and Ranjbar, Karamian and Hajmoradi (Citation2011). The mitotic chromosome number of an accession of Nisyros (Greece) of Trigonella balansae Boiss. & Reuter, an annual pasture legume of Eurasian origin, was first reported by Kamari and Papatsou (Citation1973). Studies on the impact of karyotypic data on the interspecific and phylogenetic relationships and also on meiotic behavior in the genus are still limited.
As little information exists for this genus, the present work aimed at increasing the knowledge of its cytogenetic and biogeography, reporting the base chromosome numbers and polyploidy levels between different species and comparing with previous results, which are reported throughout the world.
In this contribution, we reported the somatic chromosome numbers of 22 taxa of Trigonella belonging to eight sections. Also, chromosome numbers are determined for the first time in seven taxa of Trigonella.
Material and methods
Description of database
Some chromosome records from online databases (http://www.tropics.org/Project/IPCN) and the literature are presented in Table . Each record in the database includes the following data: name of taxa as published in the original source, the standardized name (authorship of the name is corrected, as well as possible typing errors, and the currently accepted name); data on chromosomes includes chromosome number (2n), basic chromosome number (x), the name of person who performed the chromosome count, and the locality where plant materials were collected.
Table 1. Trigonella species analyzed in the present study.
Geographical distribution of the taxa belonging to the genus Trigonella was obtained from online databases (http://ww2.bgbm.org/euroPlusMed/QUERY.asp and http://www.ildis.org/) and atlases mainly based on their localities taken from Floras and literatures and mapped here.
Cytogenetic observation
Chromosome number were analyzed in the Trigonella genus by mitotic and meiosis. For meiosis study, 15 flower buds from at least five plants at an appropriate stage of development were fixed in 96% ethanol, chloroform and propionic acid (6:3:2) for 24 h at room temperature and then stored in 70% ethanol at 4°C until used. Anthers were squashed and stained with 2% acetocarmine. Chromosome numbers were determined during diakinesis stage. The meiotic chromosome numbers were studied in 16 populations of Trigonella disperma Bornm. ex Vassilcz., eight populations of T. monantha C. A. Mey., five populations from each of T. latialata (Bornm.) Vassilcz., T. aphanoneura Rech. f. and T. coerulescens (M. Bieb.) Halácsy, four populations of T. spruneriana Boiss., three populations of T. persica Boiss., two populations from each of T. teheranica Bornm. and T. subenervis Rech. f. and only one population from each of T. adscendens Aphan. & Gontsch, T. yasujensis Ranjbar, Hajmoradi & Karamian, T. arcuata C.A. Mey. and T. asteroites Fisch. & C. A. Mey. For the mitotic study, the root tip pretreatment was conducted in 8-hydroxyquinoline for 3–4 hours; afterwards they were fixed in Farmer’s fluid (ethanol:acetic acid 3:1) for 24 h (Fukui and Nakayama Citation1996); the hydrolysis was done in Farmer’s fluid:chloridric acid 1:1 for 20–30 min at room temperature, and staining was with 2% aceto-orcein for 2 h. All slides were made permanent by venetian turpentine. Photographs of chromosomes were taken on an Olympus BX-51 (Tokyo, Japan) photomicroscope at initial magnification of ×1000. Chromosome counts were made from well-spread metaphases in intact cells, by direct observation and from photomicrographs. In five populations of T. monantha, and only one population from each of T. arcuata, T. asteroites, T. spruneriana, T. callicaeras Fischer ex M. Bieb., T. turkmena M. Popov, T. filipes Boiss., T. monspeliaca L., T. stellata Forssk. and T. orthoceras Kar. & Kir., chromosome number were counted by mitosis.
Result and discussion
Chromosome number
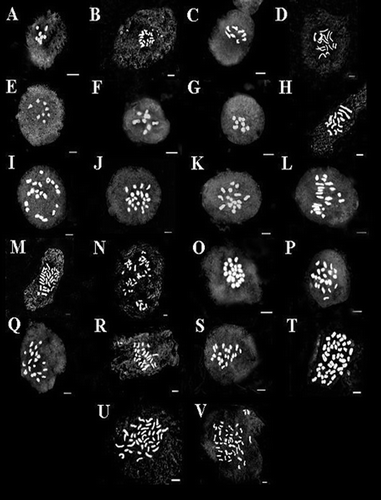
The chromosome number was studied in 58 populations of 19 species (22 taxa including subspecies). Voucher specimen related to studied species is illustrated in Table . Twelve species were annual and belong to seven different sections. In the Bucerates section, Trigonella arcuate, T. astroites and T. persica were diploid and showed chromosome number 2n = 2x = 16 (Figures A–H) while T. monantha and T. orthoceras were polyploid. 13 populations of T. monantha including four subspecies were studied. All populations of these subspecies were hexaploid with chromosome number 2n = 6x = 42 (Figure ), but one population of T. monantha ssp. monantha C. A. Mey. (Figure M) and one population of T. monantha ssp. noeana (Boiss.) Hub.-Mor. (Figure R) showed chromosome number 2n = 4x = 28 and were tetraploid. Based on these results, the base number of T. monantha is x = 7. T. orthoceras is also polyploid with chromosome number 2n = 6x = 48, which is consistent with the proposed base number of x = 8. Two pairs of satellite chromosomes were observed in this species, but none of the other members of the section (Figure V).
Table 2. Trigonella species analyzed in this study with their respective chromosome numbers, locations, and voucher specimens.
Figure 1. (A) meiosis in T. arcuata (26019); (B) mitosis in T. arcuata (23502); (C) meiosis in T. astroites (23549); (D) meiosis in T. astroites (27616); (E–G) meiosis in T. persica: (E) T. persica (23568); (F) T. persica (23706); (G) T. persica (26394); (H) mitosis in T. persica (33927); (I–M) meiosis in T. monantha ssp. monantha: (I) T. monantha ssp. monantha (25671); (J) T. monantha ssp. monantha (23700); (K) T. monantha ssp. monantha (23694); (L) T. monantha ssp. monantha (23567); (M, N) mitosis in T. monantha ssp. monantha: (M) T. monantha ssp. monantha (23513); (N) T. monantha ssp. monantha (23638); (O–Q) meiosis in T. monantha ssp. noeana: (O) T. monantha ssp. noeana (23527); (P) T. monantha ssp. noeana (23547); (Q) T. monantha ssp. noeana (23509); (R) mitosis in T. monantha ssp. noeana (23642); (S) meiosis in T. monantha ssp. geminiflora (23570); (T) mitosis in T. monantha ssp. geminiflora (23519); (U) mitosis in T. monantha ssp. incisa (23646); (V) mitosis in T. orthoceras (23610). Scale bars for meiosis figures: 5 μm and for mitotic figures: 2 μm.
Trigonella spruneriana and T. filipes from T. sect. cylindricae (Boiss.) Sirj. were diploid and had chromosome number 2n = 2x = 16 (Figures A, B). The T. sect. Reflexae (Sirj.) Vassilcz. is represented in Iran by T. monspeliaca. The diploid chromosome number for this species is 2n = 16 (Figure C). T. calliceras from T. sect. callicerates (Boiss.) Sirj. was diploid with chromosome number 2n = 2x = 16 (Figure D). The T. sect. Biebersteinianae (Sirj.) Grossh. is represented in Iran by T. coerulescenc. Five populations of T. coerulescenc have been collected from different localities of Iran, and the somatic chromosome number has been determined as 2n = 16 for all studied populations (Figures E–I). T. stellata belonging to T. sect. Falcatulae (Boiss.) Sirj. was diploid with chromosome number 2n = 2x = 16 (Figure J). T. turkmena from T. sect. Verae Sirj. was another diploid species and showed chromosome number 2n = 2x = 16 (Figure K). Chromosome number of this species is determined for the first time.
Figure 2. (A) mitosis in T. spruneriana (23518); (B) mitosis in T. filipes (8855); (C) mitosis in T. monspeliaca (5256); (D) mitosis in T. calliceras (29426); (E–I): meiosis in T. coerulescens: (E) T. coerulescens (23663); (F) T. coerulescens (33921); (G) T. coerulescens (23675); (H) T. coerulescens (25669); (I) T. coerulescens (23600); (J) mitosis in T. stellata (33788); (K) mitosis in T. turkmena (27611); (L–S) meiosis in T. disperma: (L) T. disperma (23504); (M) T. disperma (23623); (N) T. disperma (23514); (O) T. disperma (24747); (P) T. disperma (24813); (Q) T. disperma (24868); (R) T. disperma (33800); (S) T. disperma (26855); (T) meiosis in T. latialata (24488). Scale bars for meiosis figures: 5 μm and for mitotic figures: 2 μm.
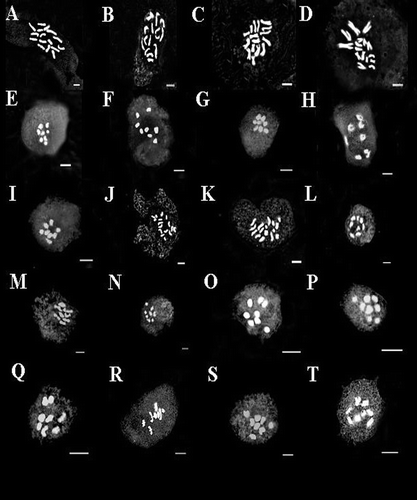
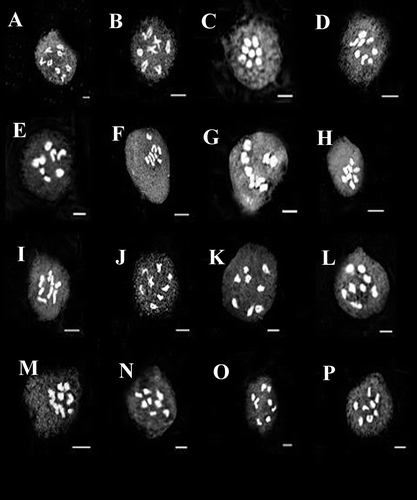
Seven species (T. disperma, T. teheranica, T. latialata, T. aphanoneura, T. adscendens, T. subenervis and T. yasujensis) were perennial and belong to T. sect. Ellipticae. Chromosome reports for all species of this section except T. disperma and T. yasujensis are presented for the first time. All studied populations of perennial species were diploid and showed chromosome number 2n = 2x = 16, which is consistent with the proposed base number of x = 8 (Figures L–T and ).
Figure 3. (A–D) meiosis in T. latialata: (A) T. latialata (23541); (B) T. latialata (29294); (C) T. latialata (23523); (D) T. latialata (24177); (E–I) meiosis in T. aphanoneura: (E) T. aphanoneura (7229); (F) T. aphanoneura (33924); (G) T. aphanoneura (33925); (H) T. aphanoneura (24675); (I) T. aphanoneura (31525); (J, K) meiosis in T. yasujensis: (J) T. yasujensis (23908); (K) T. yasujensis (33784); (L, M) meiosis in T. teheranica: (L) T. teheranica (23540); (M) T. teheranica (29477); (N, O) meiosis in T. subenervis: (N) T. subenervis (23540); (O) T. subenervis (29477); (P) T. adscendens (23599). Scale bars for meiosis figures: 5 μm and for mitotic figures: 2 μm.
Chromosome data analysis and biogeography
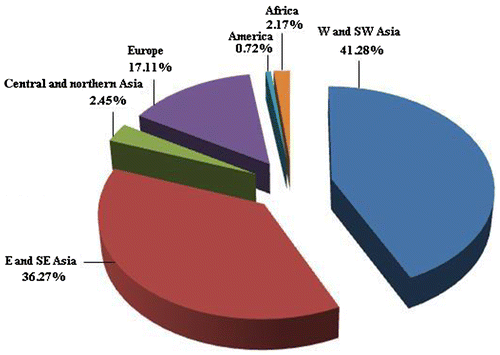
The current databases represent the reports for 70 species (93 taxa including subspecies) from total of 140 species that are estimated for the genus Trigonella throughout the world (Table ). There are some records for approximately 50% of species with known chromosome numbers. Asian taxa are more intensively studied rather than the taxa growing in other geographical areas. About 107 species of the genus occur in Asia (Boissier Citation1872; Grossheim Citation1945; Huber-Morath Citation1969; Townsend Citation1974; Rechinger Citation1984), as much as 80% of the chromosome number records originate from this continent. Even within Asia the data are not equally distributed: 41.28% of the Asian records originate from west and southwest Asia, 36.27% in east and southeast Asia and 2.45% in central and northern Asia (Figure ). This is somewhat proportional to the number of species. While 68 species occur in west and southwest Asia, 22 are in east and southeast Asia and 56 are in central and northern Asia; and of about 28 taxa in Europe, 19 taxa in Africa, and three taxa in North America that represented only 17.11%, 2.17% and 0.72% chromosome counts, respectively (Figure ). There are only two species in Australia and there are no chromosome reports from this country. Thus, an unequal spreading record throughout the distribution area of the genus is apparent. However, the hypothesis is needed to confirm by deeper karyogeographical explorations.
Another comparison is the percentage of taxa that are entirely diploid, those are both diploid and polyploid, and those are entirely polyploid. For the genus Trigonella, we have found 87.14% diploid, 7.14% diploid and polyploid, and 5.71% entirely polyploid taxa. Polyploidy variations from diploid (2n = 2x = 14) to hexaploid (2n = 6x = 48) have been reported in Trigonella species.
The basic chromosome numbers were determined as x = 7 and x = 8 for the genus Trigonella. Chromosome data from Portugal, Turkey and Iran show a base number of x = 7. However the most frequent base number is x = 8 (Tables and ). All perennial species have x = 8, while annual species have x = 7, 8. So it is clear that chromosome base number x = 8 is primary and x = 7 is secondary chromosome base number in the genus Trigonella. Hence the process of evolution in chromosome number appears to be from x = 8 to x = 7 and x = 7 might have originated from it as a result of progressive evolution as descending aneuploidy or dysploidy. In Iran most species have base number x = 8 (90%) and x = 7 occurred less frequently (10%).
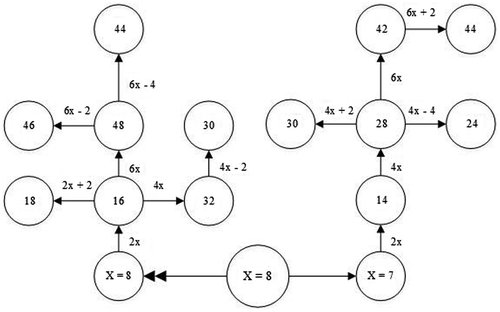
Based on the literature, a variety of chromosome numbers (2n = 14, 16, 24, 28, 30, 32, 42, 44, 46 and 48) have been reported for the genus Trigonella. Considering that the basic chromosome number is x = 7, 8, these variations in chromosome number might be related to evolutionary processes such as ascending or descending aneuploidy. Evolutionary pathways in the Trigonella genus are shown in Figure .
The chromosome numbers reported here agree with those reported in the literature in most taxa but there are some inconsistent results. Martin et al. (Citation2008) reported 2n = 16, 46 and Astanova (Citation1981) reported 2n = 44 for T. orthoceras, however we determined 2n = 6x = 48 for this species. Whereas x = 8 is reported for this species, 2n = 44 and 2n = 46 can originate from 2n = 6x = 48 as a result of descending (2n = 6x – 4 = 44) and ascending (2n = 6x + 2 = 46) aneuploidy (Figure ). We studied four populations of T. orthoceras and it was clear that all of them were polyploid, but chromosome counting was possible only in one population which reported here.
The chromosome number is determined as 2n = 28, 42 for T. monantha ssp. monantha in this study, but the somatic chromosome number has been reported as 2n = 16, 28, 30 for this taxon by Martin et al. (Citation2008). Kumari and Bir (Citation1979) reported 2n = 44 and Khatoon and Ali (Citation1991) reported 2n = 48 for T. monantha ssp. incisa (Benth.) Ali.
Based on our study, T. monantha ssp. noeana has 2n = 28, 42, but three different chromosome numbers have been reported as 2n = 14, 30, 44 for this taxon in the literature (Martin et al. Citation2008; Kliphuis and Barkoudah Citation1977; Singh and Singh Citation1976). According to Martin et al. (Citation2008), there are 4x and 6x cytotypes in almost all polyploid subspecies. The presence of polyploidy in the same populations or within one (mixopolyploidy) indicates the autopolyploid origin of the tetraploid cytotypes and suggests recurrent origins.
Based on the literature, x = 7, 8 is reported for different subspecies of T. monantha, so 2n = 30 may originate from 2n = 4x = 32 by descending aneuploidy (2n = 4x – 2 = 30) or from 2n = 4x = 28 by ascending aneuploidy (2n = 4x +2 = 30). Also 2n = 44 in these taxa can be formed by descending aneuploidy (2n = 6x – 4) from 2n = 6x = 42 or ascending aneuploidy (2n = 6x + 2) from 2n = 6x = 48 (Figure ).
We determined 2n = 16 for T. stellata but Bidak and Brandham (Citation1995) reported 2n = 18 for this species. These two different results may be related to the B chromosome or occurrence of ascending aneuploidy (2n = 2x + 2 = 18) (Figure ).
It seems these evolutionary comments and diagram could solve many complexities in chromosome numbers of the genus. In their studies on the genera Astragalus L., Salvia L., Onobrychis Mill., and Scutellaria L., Ranjbar, Hajmoradi, et al. (Citation2012), Ranjbar and Mahmoudi (Citation2013), Ranjbar and Mahmoudian (Citation2015), and Ranjbar et al. (Citation2015) also found many complexities in chromosome numbers based on evolutionary relationships.
Center of origin or diversity for Trigonella
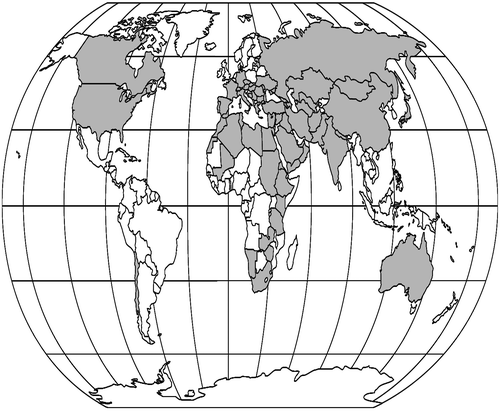
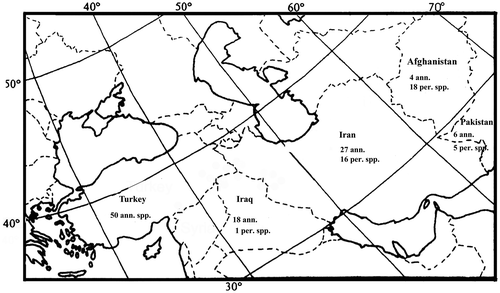
The place of origin for a species, as explained by Vavilov (Citation1951), is likely to be an area that contains a large amount of genetic variability. According to him, variation is a function of time; hence the region containing the greatest variation in a species would have supported and sustained that species for a longer time than the other regions. Vavilov defined eight geographic centers, two of which were the Near Eastern and Mediterranean centers, extending into Iran (Vavilov Citation1951). It has been postulated by Ranjbar, Karamian, et al. (Citation2012) that southwest Asia (mainly Afghanistan, Iran and Turkey) is the center of origin for Trigonella. The distribution map of Trigonella genus is given in Figure . All members of the T. sect. Ellipticae are Irano-Turanian elements and distributed only in west and southwest Asia (17 species), central and northern Asia (26 species) and east and southeast Asia (eight species). It seems that local endemism plays an important role in this section. All perennial species belong to Ellipticae section. They prefer mountainous regions with dry and windy conditions and do not form dense populations. They are found from the sea level in the region of Flora Iranica. Trigonella latialata is the only species which occurs at elevations higher than 2500 m asl (up to 3000 m asl), while the most favorable altitudes for T. sect. Ellipticae growth in Iran is between 700 and 2200 asl. This section with 13 distinct species is distributed in the central part of Iran surrounded by the central desert, around Isfahan, Yazd, Kerman, Hamadan, Kermanshah and Kurdistan Provinces. This area extends further along the northeastern Alborz in Khorasan Province to 2400 m asl, and penetrates to even higher elevations in Afghanistan. Afghanistan is one of the most important centers of diversity of the genus in the old world. Trigonella bactriana Vassilcz., T. heratensis Rech. f., T. gharuensis Rech. f. and T. xeromorpha Rech. f. are the species which occur only at elevations lower than 2200 m asl, while the most favorable altitudes in Afghanistan for the growth of T. sect. Ellipticae are between 2300 and 4000 m asl. This section with 18 distinctive species is distributed at higher elevations in Afghanistan.
Trigonella genus has 18 perennial and four annual species in Afghanistan, Pakistan has five perennial and six annual species. Turkey has 50, all of which are annual, and Iraq has one perennial and 18 annual species. Iran has 27 annual and 16 perennial species. So Turkey, Iran and Afghanistan are the diversity centers for the genus Trigonella (Figure ). It seems that over time perennial and annual species respectively penetrate to eastern and western borders from Iran. This theory is verified by results obtained from chromosome and geographical data. All perennial species are diploid, while annual species are diploid and polyploid. As polyploidy is a new phenomenon, so it is clear that annual species have evolved later. Whereas only annual species exist in the western border of Iran, Trigonella species cannot penetrate from the west to Iran. On the other hand, as explained above, all perennial species in Iran grow at low elevations but perennial species in Afghanistan prefer high elevations, and it seems that species have arisen at low elevations and gradually move toward high altitudes. Also, Iran is a significant country from the point of view of plant genetic resources and diversity. It has more than 2000 endemic plants and an immense diversity has been reported in many legumes such as Vicia L., Medicago L., Trifolium L., Lathyrus L., Onobrychis Mill., Trigonella L., Pisum L., Cicer L., and Lens Mill. Also, many genera of cultivated plants, e.g. Cicer, Lens, Pisum, Amygdalus L., Prunus L., and Triticum L. have their center of origin or diversity in this country. According to our observations all perennial Trigonella species that occur in Iran exhibited remarkable diversity. This result supports Vavilov’s (Citation1951) hypothesis. According to these comments, Trigonella species cannot penetrate from eastern borders to Iran, so Iran can be regarded as the center of origin of the Trigonella genus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding information
The fieldwork in Iran was supported by postdoctoral grants from the Bu-Ali Sina University.
Acknowledgements
The great help of Dr Vitek, Dr Wallnofer, Dr Till, Dr Sida, Dr Gautier, and Mr Fumeaux during our visit from Herbaria W, WU and PR in Vienna and Praha and G in Geneve is much appreciated. We would like to thank the director of the herbarium of Ferdowsi University of Mashhad for making the herbarium facilities available for our study for the processing and loan of herbarium specimens.
References
- Agarwal K, Gupta PK. 1983. Cytological studies in the genus Trigonella Linn. Cytologia. 48(4):771–779.
- Ahmad F, Acharya SN, Mir Z, Mir PS. 1999. Localization and activity of rRNA genes on fenugreek (Trigonella foenum-graecum L.) chromosomes by fluorescent in situ hybridization and silver staining. Theo App Gen. 98(2):179–185.
- Astanova SB. 1981. Chromosome numbers of Leguminosae of flora in Tajikistan. Dokl Akad Nauk Tadziksk SSR. 24:61–63 (In Russian).
- Badrzadeh M, Ghafarzadeh-Namazi L. 2009. Trigonella caerulea (Fabaceae), an aromatic plant from Ardebil province. Iran. Iran J Bot. 15(1):82–84.
- Bairiganjan GC, Patnaik SN. 1989. Chromosomal evolution in Fabaceae. Cytologia. 54(1):51–64.
- Balodi B, Rao RR. 1991. The genus Trigonella L. (Fabaceae) in the northwest Himalaya. J Econ Tax But. 5(1):11–16.
- Bartolo G, Brullo S, Pavone P. 1981. Números cromosomáticos de plantas occidentales. Anales Jard Bot. 38(1):288–299.
- Bhattacharya NK. 1974. Demonstration of the effect of chemicals on a few genera yielding spices of commerce. Bull Bot Soc Bengal. 28:19–24.
- Bhatti M, Khan M, Ahmad B, Jamshaid M, Ahmad W. 1996. Antibacterial activity of Trigonella foenum-graecum seeds. Fritoletapia. 67:372–374.
- Bhaumik GH. 1976. Cytological investigations on some members of the tribe Trifolieae (family Papilionaceae). Sci Cult. 42:322–324.
- Bidak L, Amin AW. 1996. Inter-and intraspecific chromosomal variations in four species of Trigonella L. J Union Arab Biol. 3(1):203–215.
- Bidak L, Brandham PE. 1995. Intraspecific uniformity of chromosome number and nuclear DNA quantity in two Egyptian weedy species, Malva parviflora (Malvaceae) and Trigonella stellata (Leguminosae). Kew Bull. 50:595–599.
- Bir SS, Kumari S. 1979. Cytological evolution of the leguminous flora of the Punjab plain. Recent Res Pl Sci. 7:252–260.
- Boissier PE. 1872. Trigonella L. In: Boissier PE, editor. Flora Orientalis. Geneva. p. 65–91.
- Chopra RN, Nayyarand SL, Chorpa IE. 1956. Glossary of Indian Medicinal Plants. 1st ed. New Delhi: CSIRO.
- Dangi RS, Lagu MD, Choudhary LB, Ranjekar PK, Gupta VS. 2004. Assessment of genetic diversity in Trigonella foenum–graecum and Trigonella caerulea using ISSR and RAPD markers. BMC Plant Biol. 4:13–23.
- Danin A, Small E. 1989. Contributions to the flora of Palestine and Sinai. V. Trigonella sibthorpii Boiss., a new record from Israel. Israel J Bot. 38:121–124.
- Darlington CD, Wylie AP. 1955. Chromosome atlas of flowering plants. London: Allen and Unwin Press.
- Das AB, Mohanty S, Das P. 2001. Cytophotometric estimation of 4C DNA content and karyotype analysis in ten cultivars of Trigonella foenum-graecum. Iran J Bot. 9(1):1–9.
- Das AB, Mohanty S, Das P. 2002. Cytophotometric estimation of 4C DNA and karyotype analysis in ten cultivars of Trigonella foenum-graecum – II. Iran J Bot. 9(2):151–159.
- Das AB, Pattnaik M, Thangaraj T, Das P. 1997. Cytophotometric estimation of nuclear DNA content and karyotype analysis of eight cultivars of Trigonella foenum-graecum. Cytobios. 91:171–179.
- Díaz Lifante Z, Luque T, Bárbara CS. 1992. Chromosome numbers of plants collected during Iter Mediterraneum II in Israel. Bocconea. 3:229–250.
- Dundas IS, Nair RM, Verlin DC. 2006. First report of meiotic chromosome number and karyotype analysis of an accession of Trigonella balansae (Leguminosae). New Zeal J Agri Res. 49(1):55–58.
- Fernandes A, Queiros M. 1978. Contribution à la connaissance cytotaxinomique des Spermatophyta du Portugal. IV. Leguminosee (Suppl. 3). Bol Soc Brot. 52:79–164.
- Fernandes A, Santos MF, Queiros M. 1977. Contribution à la connaissance cytotaxonomique des Spermatophyta de Portugal. Leguminosae. Bol Soc Brot. 51:137–186.
- Fukui K, Nakayama S. 1996. Analysis of chromosome information. In: Plant Chromosomes: Laboratory methods. Boca Raton: CRC Press; p. 241–255.
- Ghosh AK. 1980. Chromosome number of Trigonella spinosa. Curr Sci. 49:154–155.
- Girardon P, Sauvaire Y, Baccou UC, Bessiere JM. 1989. Identification de la 3-hydroxy-4, 5-dimethyl-2 (SH)-furanone dans L’arome des grains de Fenugrec (Trigonella foenum-graecum L.). Lebensm Wiss Technol. 19:44–46.
- Grossheim AA. 1945. Trigonella L. In: Shishkin BK, editor. Flora USSR. Moscow: Leningrad. p. 102–129.
- Hamzeh’ee B. 2000. Some new and noteworthy plant records from Iran. Iran J Bot. 8(2):271–277.
- Hesamzadeh, SM, Ziaei Nasab, M 2009. Color chromosome atlas of legumes collected in the natural resources gene bank of Iran. Iran: Tehran. p. 104.
- Huber-Morath A. 1969. Trigonella. In: Davis PH, editor. Flora of Turkey. Edinburgh, UK: Edinburgh University Press; p. 452–482.
- Jahan B, Vahidy AA, Ali SI. 1994. Chromosome numbers in some taxa of Fabaceae mostly native to Pakistan. Ann MO Bot Gard. 81(4):792–799.
- Jain SJ, Raghuvanshi SS. 1974. Attention to some accessories. In: Kachroo P, editor. Advancing Frontiers in Cytogenetics. Delhi: Hindustan Publ; p. 344–347.
- Janighorban M. 2004. A new record of the genus Trigonella (Fabaceae) for the flora of Iran. Iran J Bot. 10(2):177–179.
- Kamari G, Papatsou S. 1973. Chromosome studies in some Mediterranean angiosperms. Nord J Bot. 126:266–268.
- Kar K, Sen S. 1991. A comparative karyological study of root and embryo tissue of a few genera of Leguminosae. Cytologia. 56(3):403–408.
- Kesarwani R, Raghuvanshi SS. 1988. Comparison of B carrier and non-carrier populations of diploid and autotetraploid Trigonella foenum-graecum. New Bot. 15:19–22.
- Kesarwani R, Raghuvanshi SS. 1990. Frequency of B-chromosomes in C16 reverted-diploid of Trigonella foenum-graecum. J Cytol Genet. 25:24–28.
- Khatoon S, Ali SI. 1991. Chromosome numbers in subfamily Papilionoideae (Leguminosae) from Pakistan. Willdenowia. 20(1/2):159–165.
- Kirschner J, Stepanek J. 1992. In: Měsíček J, Javůrková-Jarolímová V, editors. List of chromosome numbers of the Czech vascular plants. Praha: Academia. p. 144.
- Kliphuis E, Barkoudah YI. 1977. Chromosome numbers in some Syrian angiosperms. Acta Bot Neerl. 26(3):239–249.
- Kumari S, Bir SS. 1990. Karyomorphological evolution in Papilionaceae. J Cytol Genet. 25(1):173–219.
- Ladizinsky G, Vosa CG. 1986. Karyotype and C-banding in Trigonella section Foenum-graecum (Fabaceae). Pl Syst Evol. 153(1/2):1–5.
- Laxmi V, Datta SK. 1986. Induction and analysis of MMS induced mutant of fenugreek. Sci Cult. 52:310–312.
- Laxmi V, Gupta MN, Datta SK. 1983. Investigations on an induced green seed coat colour mutant of Trigonella foenum-graecum L. Cytologia. 48(2):373–378.
- Luque T, Lifante ZD. 1991. Chromosome numbers of plants collected during Iter Mediterraneum I in the SE of Spain. Bocconea. 1:303–364.
- Mabberley DJ. 1997. The plant book, a portable dictionary of the vascular plants. 2nd ed. Cambridge: Cambridge University Press.
- Magulaev AY. 2001. On the karyology of Trigonella of Caucasus. In 3D International Conference of Biodiversity of Caucasus. Nalchik. p. 32.
- Malla SB, Saiju H, Gorkhali M, Singh MP. 1977. In IOPB chromosome number reports VI. Taxon. 26(2/3):257–274.
- Martin E, Akan H, Ekici M, Aytaç Z. 2008. Karyomorphological studies on section Bucerates Boiss. of Trigonella L. (Leguminosae) from Turkey. Caryologia. 61(2):225–236.
- Martin E, Akan H, Ekici M, Aytac Z. 2010. Karyology of ten Turkish Trigonella L. (Leguminosae) species from section Cylindricae Boiss. Turk J Bot. 34(6):485–494.
- Martin E, Akan H, Ekici M, Aytac Z. 2011a. New chromosome numbers in the genus Trigonella L. (Fabaceae) from Turkey. Afr J Biotechnol. 10(2):116–125.
- Martin E, Akan H, Ekici M, Aytac Z. 2011b. Karyotype analyses of ten sections of Trigonella (Fabaceae). Comp Cytogenet. 5(2):105–121.
- Murin A, Ferakova V. 1981. Caryological study of Slovakian flora III. Acta Fac Rerum Nat Univ Comenianae Bot. 28:59–62.
- Pavlova D. 1996. Mediterranean chromosome number reports. Fl Medit. 6:323–328.
- Qi Rh. 1997. A comparative study on three species of Trigonella L. Grassl China. 4:52–54.
- Raghuvanshi SS, Mahajan S. 1982. Chromosomal associations in B carrier and noncarrier diploid, tetraploid and octoploid Impatiens balsamina L. Proc Indian Acad Sci. 48(1):147–151.
- Raghuvanshi SS, Upreti M. 1977. A new category of B-chromosomes in Trigonella foenum-graecum L. Curr Sci. 46(22):789–790.
- Rakhee SD, Meena DL, Lal BC, Prabhakar KR, Vıdya SG. 2004. Assessment of genetic diversity in Trigonella foenum-graecum and Trigonella caerulea using ISSR and RAPD markers. BMC Plant Biol. 4:1–11.
- Ranjbar M, Hajmoradi Z. 2012. Notes on Medicago sect. Lunatae Boiss. and Trigonella sect. Bucerates Boiss. of the tribe Trifolieae (Fabaceae), with two new records from Iran. Iran J Bot. 18(2):235–238.
- Ranjbar M, Hajmoradi Z. 2015. A new species of Trigonella sect. Ellipticae (Leguminosae-Papilionoideae) from Iran, including cytogenetic and anatomical notes. Phytotaxa. 202(1):26–34.
- Ranjbar M, Hajmoradi Z. 2016. Comparative leaf epidermis and anatomical study in populations of Trigonella spruneriana (Fabaceae) from Iran. Webbia. 71(1):81–89.
- Ranjbar M, Hajmoradi Z, Karamian R. 2011. Cytogenetic study and pollen viability of four populations of Trigonella spruneriana Boiss. (Fabaceae) in Iran. J Cell Mol Res. 3(1):19–24.
- Ranjbar M, Hajmoradi Z, Karamian R. 2014. Novelty in Trigonella sect. Ellipticae (Fabaceae) from. Iran. 23(2):209–216.
- Ranjbar M, Hajmoradi F, Karamian R. 2012. An overview oncytogenetics of the genus Onobrychis (Fabaceae) with special reference to O. sect. Hymenobrychis from Iran. Caryologia. 65(3):187–198.
- Ranjbar M, Karamian R, Hajmoradi Z. 2011. Cytomorphological study of Trigonella disperma (Fabaceae) in Iran. Cytologia. 76(3):279–294.
- Ranjbar M, Karamian R, Hajmoradi Z. 2012. A new species and taxonomy studies in Trigonella sect. Ellipticae (Fabaceae) in Iran. Ann Bot Fen. 49(4):279–287.
- Ranjbar M, Karamian R, Hajmoradi Z, Joharchi MR. 2012. A revision of Trigonella sect. Ellipticae (Fabaceae) in Iran. Nord J Bot. 30(1):17–35.
- Ranjbar M, Mahmoudi C. 2013. Chromosome numbers and biogeography of the genus Scutellaria L. (Lamiaceae). Caryologia. 66(3):205–214.
- Ranjbar M, Mahmoudian B. 2015. An overview on cytogenetics of the genus Astragalus subgenus Hypoglottis (Fabaceae). Caryologia. 68(2):109–124.
- Ranjbar M, Pakatchi A, Babataheri Z. 2015. Chromosome number evolution, biogeography and phylogenetic relationships in Salvia (Lamiaceae). Webbia. 70(2):293–312.
- Rechinger KH. 1984. Trigonella. In: Rechinger KH editor. Flora Iranica, Vol. 157. Graz-Austria: Akademische Druck- und Verlagsanstalt. p. 207–253.
- Runemark H. 2006. Mediterranean chromosome number reports. Fl Medit. 16:408–425.
- Saggoo MIS, Kaur J. 1989. SOCGI plant chromosome number reports. J Cytol Genet. 24:179–183.
- Sanjappa M, Dasgupta A. 1977. In IOPB chromosome number reports LVI. Taxon. 26(2/3):257–274.
- Sareen TS, Singh PD. 1976. Cytological studies in some north Indian Leguminosae. Proc Indian Sci Congr Assoc. 63:122–123.
- Sharma AK. 1970. Annual report. Res Bull Univ. 2:1–50.
- Singh A. 1973. Studies on the interspecific hybrids of Trigonella corniculata L., T. hamosa L. and T. cretica L. Genetica. 44(2):264–269.
- Singh A, Roy RP. 1970. Karyological studies in Trigonella. Indigofera and Phaseolus. The Nucleus. 13(1):41–54.
- Singh A, Singh D. 1976. Karyotype studies in Trigonella. Nucleus. 19:13–16.
- Slavĭk B, Jarolĭmovă V, Chrtek J. 1993. Chromosome counts of some plants from Cyprus. Candollea. 48(1):221–230.
- Townsend CC. 1974. Trigonella L. In: Townsend CC, Guest E, editors. Flora of Iraq. Baghdad: Min Agric; p. 86–111.
- Vavilov NI. 1951. The origin, variation, immunity and breeding of cultivated plants. Chron Bot. 13(1/6):1–366.
- Yan GX, Zhang SZ, Xue FH, Wang LY, Yun JF, Fu XQ. 2000. The chromosome numbers and natural distribution of 38 forage plants in north China. Grassl China. 5:1–5.
- Yeh MS, Yuasa H, Maekawa F. 1986. Chromosome numbers in the Leguminosae. Sci Rep Res Inst Evol Biol. 3:57–71.
- Yilmaz A, Martin E, Unal F, Akan H. 2009. Karyological study on six Trigonella L. species (Leguminosae) in Turkey. Caryologia. 6(2):89–94.