Abstract
The knowledge about the genome of the burbot Lota lota (Linnaeus, 1758), the only freshwater gadiform fish (Gadidae, Gadiformes), is limited to chromosome number (2n = 48, NF = 78). In this work analysis of burbot chromosomes by C-banding, DAPI staining and restriction endonuclease digestion proved both heterogeneity of the heterochromatin and interchromosomal differences in the vulnerability to the endonuclease treatment. Silver nitrate and chromomycine A3 staining and fluorescence in situ hybridization using 28S rDNA probes showed that nucleolus organizer regions appear in one pair of burbot chromosomes. There is also a single locus for 5S rDNA repeats exhibited in burbot; however, major and minor rRNA gene families are not linked. The single and separated location of major and minor rDNA families suggests that the burbot karyotype still retains some characteristics of the putative ancestral karyotype. Although the increased number of the chromosome arms in relation to the ancestral karyotype suggests that burbot karyotype experienced several inversions, we did not find any intercalary telomeric DNA sequences in their chromosomes.
1. Introduction
Gadiformes (Pisces, Actinopterygii, Teleostei) comprises more than five hundred species belonging to 75 genera of nine families (Nelson Citation2006). Genetic studies of gadiforms are usually restricted to species important in fisheries (cods, haddocks, hakes, whitings) and are limited to evaluation of genetic variation and differentiation among populations. From the cytogenetic point of view, this group has been rarely studied and karyological data have been provided only for 16 species (Arai Citation2011; Ghigliotti et al. Citation2012; Garcia-Souto et al. Citation2015), mostly describing chromosome numbers.
The burbot (Lota lota) is the only freshwater gadiform species. Fossil evidence and genetic data suggest that the freshwater form of Lota appeared in Europe 15–5 Ma, colonizing North America afterwards (Van Houdt et al. Citation2003). Adaptation to the freshwater environment and further evolution under the new conditions may be associated with changes in the burbot genome, including chromosomal rearrangements and mutations (Jones et al. Citation2012), therefore making the cytogenetic analysis of this species particularly interesting.
In this research we applied classical and molecular cytogenetic techniques to study the chromosomal distribution of heterochromatic and GC- and AT-rich regions, locate the nucleolar organizer regions (NORs) and to physically map rRNA genes and telomeric DNA sequences in metaphase chromosomes of the burbot. Results were discussed in comparison with the available cytogenetic data from other gadiform fishes.
2. Materials and methods
2.1. Fish sampling
Twelve one-year-old specimens of burbot (nine females and three males) were obtained from the fish hatchery of the Department of Riverine and Lake Fisheries in Olsztyn (north-eastern Poland: 53°45′14ʺ N, 20°27′43ʺ E).
2.2. Chromosome preparation
Chromosomes were prepared from cephalic kidney cells following Jankun et al. (Citation1998). Fishes were injected with 0.1% colchicine (1 ml per 100 g of body weight) and, 60 min later, sacrificed by an overdose of the anesthetic MS-222 (Sigma-Aldrich, Saint Louis, MO, USA). Kidneys were then removed and dissected in 0.075 M KCl to obtain cell suspension that were hypotonized for 60 min in 0.075 M KCl, prefixed (5 min) and fixed (30 min) in methanol: acetic acid (3:1), washed twice in fixative, and spread onto microscope slides. Chromosome spreads were stained in buffered 4% Giemsa (pH 6.8, 10 min).
2.3. Banding techniques
Heterochromatin was identified using the C-banding technique of Sumner (Citation1972) with some modifications (Śliwińska-Jewsiewicka et al. Citation2015). Chromosome preparations were digested with AluI, DdeI or HinfI (Promega, Madison, WI, USA), which are usually applied to fish chromosomes (Table ) according to previously published methods (Jankun et al. Citation2004). AgNO3 staining of the NORs was performed according to Howell and Black (Citation1980). GC-rich chromatin was detected by chromomycin A3 (CMA3) staining (Sola et al. Citation1992) and AT-rich chromatin with 4′,6-diamidino-2-phenylindole (DAPI) (Śliwińska-Jewsiewicka et al. Citation2015).
Table 1. Restriction enzymes and chromosome digestion conditions.
2.4. FISH
rDNA sites were mapped to the chromosomes of burbot using fluorescence in situ hybridization (FISH) with 28S and 5S rDNA probes according to Fujiwara et al. (Citation1998) with a slight modification (Kirtiklis et al. Citation2014). Probes were obtained via PCR using primers described by Zardoya and Meyer (Citation1996) (F8–28S: 5′-TGAAATACCACTACTCTTATCGTT-3′; R8–28S: 5′-GGATTCTGACTTAGAGGCGTTCAG-3′) and Komiya and Takemura (Citation1979) (5S-1: 5′-TACGCCCGATCTCGTCCGATC-3′; 5S-2: 5′-CAGGCTGGTATG GCCGTAAGC-3′). 28S and 5S rDNA probes were labeled by nick translation with fluorescein-12-dUTP and tetramethylrhodamine-5-dUTP (Roche, Mannheim, Germany), respectively. Chromosome preparations pretreated with RNase (1 mg ml−1, 37 °C, 60 min) and denatured in 70% formamide (80 °C, 1–2 min) were hybridized overnight. Posthybridization washes were performed at low stringency (37 °C, 20 min). Chromosomes were counterstained with DAPI-containing Vectashield (Vector, Burlingame, CA, USA).
Telomeric sequences were detected by FISH with a fluorescein-labeled telomere-specific peptide nucleic acid (PNA) probe (DAKO, Glostrup, Denmark) according to the manufacturer’s protocol as modified by Śliwińska-Jewsiewicka et al. (Citation2015).
2.5. Microscopy and image processing
Chromosome preparations were analyzed using Zeiss Axio Imager.A1 (Goettingen, Germany) and Nikon Eclipse 90i (Tokyo, Japan) microscopes equipped with epifluorescence, appropriate filter sets for multicolor FISH and digital cameras (Applied Spectral Imaging, Galilee, Israel and Jenoptic, Jena, Germany, respectively). Metaphase plates scoring (at least 10 per individual) and picture processing was performed as previously published (Śliwińska-Jewsiewicka et al. Citation2015). To construct karyotypes, chromosomes were organized by size and centromere position.
3. Results
The diploid chromosome number of the burbot individuals under study was invariably 2n = 48. The karyotype comprised 12 metacentric (m), 18 submetacentric (sm) and 18 subtelo-acrocentric (st-a) chromosomes (NF = 78) (Figure ).
Figure 1. Giemsa-stained karyotypes of the burbot, Lota lota after C-banding (a) and digestion with the restriction endonucleases DdeI (b), AluI (c) and HinfI (d); m: metacentric chromosomes, sm: submetacentric chromosomes, st-a: subtelo-acrocentric chromosomes. Scale bar = 10 μm.
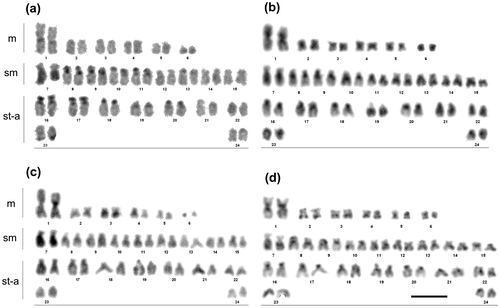
Heterochromatic C-positive bands were exhibited at the pericentromeric locations on nine chromosome pairs and in the telomeric regions on the short arms of four chromosome pairs (Table ). Moreover, the long arms of chromosome pair 7 were entirely hetrochromatinized and showed length heteromorphism in one of the specimens (Figure (a)).
Table 2. Chromosomal distribution of C-positive heterochromatin and regions resistant to restriction endonucleases in the burbot.
After treatment with DdeI, AluI and HinfI (Figure (b), (c), and (d), respectively), euchromatin was entirely digested while many heterochromatin blocks remained undigested and others were differentially digested. As summarized in Table , the C-positive heterochromatin in 7q remained undigested after DdeI, AluI and HinfI treatments whereas heterochromatin region on 6q remained undigested only after DdeI treatment. The pericentromeric heterochromatin, excluding that in pair no. 3, persisted mostly untouched after application of DdeI but was completely digested by AluI (Figure (b) and (c)). HinfI digested pericentromeric heterochromatin from three small m-sm chromosome pairs, only (Figure (d)). Furthermore, the pericentromeric chromatin on pair 19 was resistant to HinfI but completely digested by AluI and DdeI. Moreover, all the distal part of 19q was resistant to DdeI.
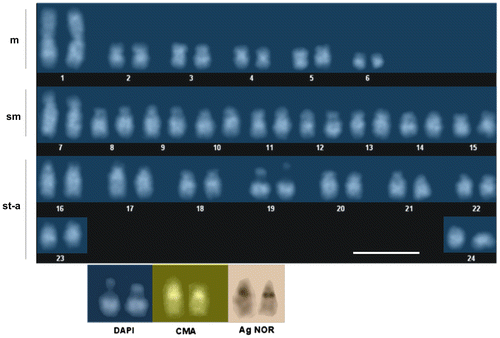
While all submetacentric pairs showed DAPI-positive regions on their long arms (Figure ), in proximal 19q appeared a DAPI-negative region that was also strongly stained by both silver and CMA3.
Figure 2. DAPI-stained karyotype of the burbot Lota lota. The bottom row shows the NOR-bearing chromosome pair (no. 19) after sequential DAPI/chromomycin A3/silver nitrate staining. Scale bar = 10 μm.
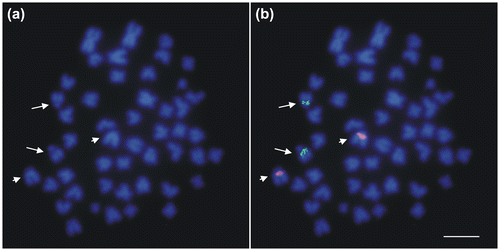
Double-color FISH experiments using 28S and 5S rDNA probes displayed clearly separated hybridization signals (Figure ). Major rDNA signals were coincident with the DAPI-negative/CMA-positive region in 19q whereas 5S rDNA signals appeared on the DAPI-positive band on 8p (Figure (a)). No variation in number, nor in intensity of the hybridization spots were detected. Moreover, no differences between males and females were found.
Figure 3. FISH mapping of 28S (arrows) and 5S (arrowheads) rDNA clusters to metaphase chromosomes of the burbot Lota lota stained with DAPI. Scale bar = 10 μm.
FISH with the PNA telomere probe revealed hybridization signals only at the ends of the sister chromatids in all burbot chromosomes (Figure ). The intensity of the signals did not differ significantly between chromosomes. Similar patterns of hybridization spots were displayed by male and female metaphase cells.
4. Discussion
Diploid chromosome numbers in Gadidae fishes vary from 2n = 26 in Eleginus species to 2n = 48 in Trispoterus minutes, Raniceps raninus and the burbot (Ráb Citation1986; Arai Citation2011; present study). In all these fishes the number of chromosome arms ranges from NF = 48 in the species with 2n = 26 to NF = 78 in the fishes with 2n = 48. Taking into consideration that a karyotype composed of 48 mono-armed chromosomes is regarded ancestral in teleostean fishes (Ohno Citation1970), gadid karyotypes must have undergone a series of rearrangements, including chromosome fusions and inversions, that in some species decreased chromosome numbers maintaining the number of chromosomal arms while in others, including burbot, increased the number of chromosome arms. As chromosomal inversions seemed to play an important role in the karyotype changes experienced by threespine sticklebacks (Gasterosteus aculeatus) during their adaptation to freshwater conditions (Jones et al. Citation2012), it is also possible that pericentric inversions were partly responsible for the increase in the number of chromosome arms found in burbot.
In fish, chromosome rearrangements may retain telomeric DNA sequences at the fusion sites (Ocalewicz Citation2013; Ocalewicz et al. Citation2013). In burbot, lack of the internally located telomeric DNA sequences suggested that breakpoints in the ancestral chromosomes were out of the telomeric area and the further chromosomal rearrangements left no telomeric repeats at the fusion sites (Nanda et al. Citation1995).
Heterochromatin, an important part of the eukaryotic genome, is composed of various types of repetitive DNA sequences and participates in nuclear organization, chromosome segregation and regulation of the gene expression (Grewal and Jia Citation2007; Varriale et al. Citation2008). So far, heterochromatin in gadid fish species has not been investigated. Our results showed varied resistance of the different blocks of burbot heterochromatin to the studied endonucleases. Interchromosomal differences in vulnerability for the endonuclease treatment could be partly explained by the diverse repetitive DNA sequence composition of the heterochromatin. In this sense, only some of the burbot chromosomes display DAPI positive (AT-rich) chromatin stained centromeric regions. Moreover, pericentromeric regions from different chromosomes showed different susceptibility for the endonuclease digestion. Such diversity confirmed that centromeric chromatin is assembled of various repetitive DNA sequences that may be interspersed with the AT-rich DNA fractions (Pidoux and Allshire Citation2005).
In contrast, CMA3-positive GC-rich chromatin in the burbot was limited to the silver nitrate stained NORs. This is coincident with the situation in other fishes in which GC-rich CMA3 positive sites usually correspond to the NORs (e.g. Ráb et al. Citation1999; Jankun et al. Citation2003, among others). NORs have been located by FISH in only other two species of gadid fishes. As happened in the burbot, the European hake Merlucius merlucius also showed a single NOR (Garcia-Souto et al. Citation2015). In contrast, the Atlantic cod (Gadus morhua) presented two different patterns, one including four NORs and the other showing six (Ghigliotti et al. Citation2012). A single chromosomal location of the NOR-related rDNA clusters is the most frequently observed pattern in fishes (Gornung Citation2013) and may be the ancestral situation. Transposition and chromosome rearrangements including breaks within the NORs may result in the redistribution and multichromosomal location of the rDNA loci (Nakajima et al. Citation2012). The presence of a single 5S rDNA cluster in burbots is coincident with the situation in many fish species and has been proposed as the ancestral condition (for a review see Martins and Wasco Citation2004). In the other gadid species studied so far a single 5S rDNA cluster has also been reported for the European hake (Garcia-Souto et al. Citation2015), but multiple clusters were observed in the Atlantic cod (Ghigliotti et al. Citation2012). The location of minor and major rDNA clusters on separate chromosomes in the burbot, as well as in the Atlantic cod and the European hake, is coincident with the situation observed in most teleost fishes (Martins Citation2007) and is regarded as the ancestral condition.
5. Conclusions
The results obtained in this work allowed a deeper characterization of the chromosomes of the burbot, the only freshwater cod-like species, showing that in spite of some changes, the karyotype of the burbot still retains many ancestral characteristics.
Ethical statement
All manipulations and the experimental procedures were provided according to the positive opinion number 27/2010 of the Local Ethical Commission from the University of Warmia and Mazury in Olsztyn, Poland.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the University of Warmia and Mazury in Olsztyn, Poland [grant number 18.610.003-300].
Acknowledgments
The authors thank the two anonymous reviewers for all valuable comments.
References
- Arai R. 2011. Fish karyotypes: a check list. Tokyo, Berlin, Heidelberg, New York: Springer
- Fujiwara A, Abe S, Yamaha E, Yamasaki F, Yoshida MC. 1998. Chromosomal localization and heterochromatin association of ribosomal RNA genes loci and silver stained nucleolar organizer regions in salmonid fishes. Chromosome Res. 6(6):463–471. 10.1023/A:1009200428369
- Garcia-Souto D, Troncoso T, Perez M, Pasantes JJ. 2015. Molecular cytogenetic analysis of the European hake Merlucius merlucius (Merlucciidae, Gadiformes): U1 and U2 snRNA gene clusters map to the same location. PLoS ONE. 10(12):e0146150. 10.1371/journal.pone.0146150
- Ghigliotti L, Fevolden S-E, Cheng C-HC, Babiak I, Dettai A, Pisano E. 2012. Karyotyping and cytogenetic mapping of Atlantic cod (Gadus morhua Linnaeus, 1758). Animal Genet. 43:746–752. 10.1111/age.2012.43.issue-6
- Gornung E. 2013. Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: A review of research. Cytogenet Genome Res. 141:90–102.
- Grewal SIS, Jia S. 2007. Heterochromatin revised. Nature Rev Genet. 8:35–46. 10.1038/nrg2008
- Howell WM, Black DA. 1980. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia. 36:1014–1015. 10.1007/BF01953855
- Jankun M, Ocalewicz K, Mochol M. 2004. Chromosome banding studies by replication and restriction enzyme treatment in vendace (Coregonus albula) (Salmonidae, Salmoniformes). Folia biologica (Krakow). 52:47–51.
- Jankun M, Ocalewicz K, Pardo BG, Martinez P, Woznicki P, Sanchez L. 2003. Chromosomal characteristics of rDNA in European grayling Thymallus thymallus (Salmonidae). Genetica. 119:219–224. 10.1023/A:1026022415908
- Jankun M, Ocalewicz K, Woznicki P. 1998. Replication, C- and fluorescent chromosome banding patterns in European whitefish, Coregonus lavaretus L. Hereditas. 128:195–199.
- Jones, FC, Grabherr, MG, Chan, YF, Russell, P, Mauceli, E, Johnson, J, Swofford, R, Pirun, M, Zody, MC, White, S, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 484:55–61. 10.1038/nature10944
- Kirtiklis L, Ocalewicz K, Wiechowska M, Boroń A, Hliwa P. 2014. Molecular cytogenetic study of the European bitterling Rhodeus amarus (Teleostei: Cyprinidae: Acheilognathinae). Genetica. 142:141–148. 10.1007/s10709-014-9761-x
- Komiya H, Takemura S. 1979. Nucleotide sequence of 5S ribosomal RNA from rainbow trout (Salmo gairdnerii) liver. J Biochem. 86:1067–1080.
- Martins C. 2007. Chromosomes and repetitive DNAs: a contribution to the knowledge of the fish genome. In: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor G, editors. Fish Cytogenetics. Enfield, Jersey, Plymouth: Science Publishers; p. 421–432.
- Martins C, Wasco AP. 2004. Organization and evolution of 5S ribosomal DNA in the fish genome. In: Williams CR, editor. Focus on genome research. New York, NY: Nova Biomedical Books; p. 335–364.
- Nakajima RT, Cabral-de-Mello DC, Valente GT, Venere PC, Martins C. 2012. Evolutionary dynamics of rRNA gene clusters in cichlid fish. BMC Evol Biol. 12:198. 10.1186/1471-2148-12-198
- Nanda I, Schneider-Rasp S, Winking H, Schmid M. 1995. Loss of telomeric sites in the chromosomes of Mus musculus domesticus (Rodentia: Muridae) during Robertsonian rearrangements. Chromosome Res. 3:399–409. 10.1007/BF00713889
- Nelson JS. 2006. Fishes of the world. 4th ed. Hoboken, New Jersey: John Wiley & Sons Inc.
- Ocalewicz K. 2013. Telomeres in fishes. Cytogenet Genome Res. 141:114–125. 10.1159/000354278
- Ocalewicz K, Furgala-Selezniow G, Szmyt M, Lisboa R, Kuciński M, Lejk AM, Jankun M. 2013. Pericentromeric location of the telomeric DNA sequences on the European grayling chromosomes. Genetica. 141:409–416. 10.1007/s10709-013-9740-7
- Ohno S. 1970. Evolution by gene duplication. New York, NY: Springer Verlag.
- Pidoux AL, Allshire RC. 2005. The role of heterochromatin in centromere function. Philosophical Transactions of the Royal Society B: Biological Sciences. 360:569–579. 10.1098/rstb.2004.1611
- Ráb P. 1986. Karyotype of the European burbot, Lota lota (L.) (Gadidae). Journal of Ichthyology. 26:127–131.
- Ráb P, Rábova M, Reed KM, Phillips RB. 1999. Chromosomal characteristics of ribosomal DNA in the primitive semionotiform fish, longnose gar Lepisosteus osseus. Chromosome Res. 7:475–480. 10.1023/A:1009202030456
- Śliwińska-Jewsiewicka A, Kuciński M, Kirtiklis L, Dobosz S, Ocalewicz K, Jankun M. 2015. Chromosomal characteristics and distribution of rDNA sequences in the brook trout Salvelinus fontinalis (Mitchill, 1814). Genetica. 143:425–432. 10.1007/s10709-015-9841-6
- Sola L, Rossi AR, Iaselli V, Rasch EM, Monaco PJ. 1992. Cytogenetics of bisexual/unisexual species of Poecilia. II. Analysis of heterochromatin and nucleolar organizer regions in Poecilia mexicana by C-banding and DAPI, quinacrine, chromomycin A3, and silver staining. Cytogenet Cell Genome. 60:229–235. 10.1159/000133346
- Sumner AT. 1972. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 75:304–306. 10.1016/0014-4827(72)90558-7
- Van Houdt JK, Hellemans B, Volckaert FAM. 2003. Phylogenetic relationships among Palearctic and Nearctic burbot (Lota lota): Pleistocene extinctions and recolonization. Mol Phylogenet Evol. 29:599–612. 10.1016/S1055-7903(03)00133-7
- Varriale A, Torelli G, Bernardi G. 2008. Compositional properties and thermal adaptation of 18S rRNA in vertebrates. RNA. 14:1492–1500. 10.1261/rna.957108
- Zardoya R, Meyer A. 1996. Evolutionary relationships of the coelacanth, lungfishes, and tetrapods based on the 28S ribosomal RNA gene. Proc Natl Acad Sci U.S.A. 93:5449–5454. 10.1073/pnas.93.11.5449