Abstract
American Foulbrood (AFB) is a particularly pernicious bacterial disease of honey bees due to the extreme persistence of endospores of the causative agent Paenibacillus larvae. These spores are resistant to harsh environmental conditions, unaffected by antimicrobial agents and can remain viable for decades. The germination of the endospore in the larval midgut is the crucial first step leading to infection, followed by vegetative growth, tissue invasion and disease, culminating in spore formation when the host´s nutrients have been exhausted. Therefore, inhibiting spore germination or impeding early vegetative growth would be a highly effective strategy for limiting the impact of AFB. We previously showed that honey bee-specific lactic acid bacteria (hbs–LAB) had a major inhibitory effect on P. larvae both in culture and in larval bioassays. The present study documents the progress towards characterization of compounds, processes and interactions between P. larvae and the hbs–LAB responsible for this inhibitory effect. Firstly, we used an agar diffusion assay and larval infection bioassay to show that most, if not all, of the inhibitory effect was associated with the extracellular fraction (secretome). Secondly, we employed a turbidimetric growth assay to demonstrate that the hbs–LAB secretome strongly inhibited P. larvae vegetative growth, however, probably not by reducing spore germination. The inhibition was similarly effective against both major P. larvae genotypes (ERIC-I and II) in all experiments. The implications of our results for characterization of the secretome and for the management and treatment of AFB and P. larvae are further discussed.
Introduction
Honey bees are important pollinators in agriculture and food production (Gallai, Salles, Settele, & Vaissière, Citation2009). Recent large-scale losses of managed honey bee colonies have led to an increased focus on honey bee health and diseases. American foulbrood (AFB) is a highly contagious and destructive bacterial honey bee brood disease affecting beekeeping worldwide (Genersch, Citation2010a). The causative agent of AFB is the spore-forming, Gram-positive bacterium Paenibacillus larvae. The infectious form of the bacterium is the extremely tenacious endospore, which enters the food canal of young honey bee larvae through feeding on spore-contaminated food (Bamrick, Citation1967; Woodrow, Citation1942). After the spore germinates, P. larvae multiplies and proliferates in the midgut, breaching the epithelium and invading the hemocoel of the honey bee larvae. The invasion coincides with the death of the infected larva, which decomposes into a brown, glue-like liquid containing vast numbers of P. larvae spores (Genersch, Citation2010a). The spores are ready to be distributed to new larvae within the colony and between colonies, either by bees (through swarming and robbing) or by beekeeping practices (Fries, Lindström, & Korpela, Citation2006; Genersch, Citation2010b). Several different P. larvae genotypes have been identified with different growth phenotypes and virulence characteristics at both the individual and colony level, and these can be distinguished using repetitive element PCR (rep-PCR) and Enterobacterial Repetitive Intergenic Consensus (ERIC) primers (Genersch et al., Citation2006; Genersch & Otte, Citation2003).
American foulbrood is a major problem for apiculture that is currently managed in most countries by quarantine and incineration of either the whole colony or just brood frames and bees and in some countries also by prophylactic treatment with common antibiotics like oxytetracycline and tylosin (Elzen et al., Citation2002; Evans, Citation2003). There is strong evidence that P. larvae is developing resistance to these antibiotics in countries where antibiotic treatment is commonly used (Elzen et al., Citation2002; Evans, Citation2003). In the European Union it is illegal to use antibiotics for treatment or prevention of AFB (Al-Waili, Salom, Al-Ghamdi, & Ansari, Citation2012; Animal and Plant Health Agency, Citation2017; Cogliani, Goossens, & Greko, Citation2011). Moreover, antimicrobial agents only affect the vegetative stage of P. larvae and not the resistant spores, which remain viable for decades, such that antibiotic treatment unfortunately has little impact on the long-term epidemiology and control of AFB (Alippi, López, Reynaldi, Grasso, & Aguilar, Citation2007; Genersch, Citation2010b).
Previous studies have looked into the potential of beneficial bacteria to control honey bee pathogens. These include trials with individual lactic acid bacteria (LAB) (Audisio et al., Citation2011; Evans & Lopez, Citation2004; Maggi et al., Citation2013) and various species of Bacillus (Benitez, Velho, de Souza da Motta, Segalin, & Brandelli, Citation2012; Minnaard & Alippi, Citation2016; Sabaté, Carrillo, & Audisio, Citation2009; Yoshiyama & Kimura, Citation2009). The health benefits to the host from beneficial microbiota can be conditioned via various mechanisms, i.e., through the enhancement of the bee’s innate immunity by LAB such as Bifidobacterium asteroides 26p, Lactobacillus kunkeei 14 and Fructobacillus fructosus 49a (Evans & Lopez, Citation2004; Forsgren, Olofsson, Vásquez, & Fries, Citation2010; McNally, Viana, & Brown, Citation2014) or by the production of antimicrobial compounds for the direct inhibition of honey bee pathogens such as P. larvae (Audisio et al., Citation2011), Melissococcus plutonius (Vásquez et al., Citation2012), and Nosema ceranae (Maggi et al., Citation2013).
However, the most interesting approach is to use the honey bee’s own honey bee specific (hbs) LAB, which includes 13 bacterial species within the Lactobacillus and Bifidobacterium genera (Forsgren et al., Citation2010; Killer, Dubná, Sedláček, Švec 2014; Vásquez et al., Citation2012). It is possible that these bacteria act as a community by cross-metabolizing each other’s secreted compounds, producing a richer diversity of metabolites and antimicrobial peptides than is possible with individually cultivated bacteria (McNally et al., Citation2014), and that may therefore be more effective, or even specifically adapted, against just honey bee pathogens. In a previous study, it was demonstrated that feeding young honey bee larvae with a mix of 11 hbs–LAB (except Lactobacillus apinorum Fhon13N and Lactobacillus mellifer Bin4N) cell suspension decreased the proportion of larvae dying from P. larvae infection (Forsgren et al., Citation2010). In this study, we followed up this work with a more detailed characterization of the nature of the inhibitory effects from the co-cultivated hbs–LAB community on P. larvae, through an analysis of the Cell-Free Supernatant (CFS), the secretome, from a mixed culture of 13 hbs–LAB species (Olofsson, Alsterfjord, Nilson, Butler, & Vásquez, 2014a; Vásquez et al., Citation2012). Our results represent a first step towards the identification and characterization of compounds involved in the inhibitory mechanisms of hbs–LAB against pathogens such as P. larvae. The long-term goal would be to use beneficial bacteria or their products as an additional and sustainable strategy for combatting AFB in honey bee colonies.
Methods
Paenibacillus larvae cultivation
Paenibacillus larvae genotypes ERIC-I (CCUG 48979) and ERIC-II (CCUG 48972) were cultured on MYPGP agar (Mueller–Hinton broth, yeast extract, potassium phosphate, glucose and pyruvate) plates as described by Nordström and Fries (Citation1995) and incubated at 35 °C with 5% CO2 for 72 h. To prepare spore suspensions with defined concentrations, P. larvae genotypes ERIC-I and ERIC-II were cultured on MYPGP agar and incubated at 35 °C with 5% CO2 for 3 weeks. Bacterial colonies were suspended in sterile 0.9% saline and a total microscopic count of the spores was made in a Helber bacteria counting chamber (Hawksley) using a phase-contrast light microscope (400×, Reichert, Austria). Stock spore suspensions were stored at 4 °C. To prepare suspensions of vegetative cells, P. larvae spores were inoculated in MYPGP broth and incubated at 35 °C with 5% CO2. After 24–30 h incubation, the samples were checked for P. larvae cells by phase contrast microscope (Forsgren, Stevanovic, & Fries, Citation2008).
Hbs–LAB cultivation
The 13 hbs–LAB (Lactobacillus kunkeei Fhon2N, Lactobacillus apinorum Fhon13N, Lactobacillus mellis Hon2N, Lactobacillus mellifer Bin4N, Lactobacillus apis Hma11N, Lactobacillus helsingborgensis Bma5N, Lactobacillus melliventris Hma8N, Lactobacillus kimbladii Hma2N, Lactobacillus kullabergensis Biut2N and Bifidobacterium asteroides Bin2N, Bifidobacterium asteroides Bin7N, Bifidobacterium asteroides Hma3N and Bifidobacterium coryneforme Bma6N) were cultured individually in supplemented MRS medium (sMRS) containing 2% fructose (Merck, Sollentuna, Sweden) and 0.1% l-cysteine (Sigma-Aldrich, Stockholm, Sweden) as previously described (Lamei et al., Citation2017). The hbs–LAB mixture used for experimentation was prepared by mixing the 13 species, each at a starting cell density of OD600=0.02, followed by incubation of the mixture in sMRS broth for 18 h at 35 °C.
The composition of the hbs–LAB mixture post-incubation was determined by cultivating bacterial colonies from the final mixture on sMRS plates, transferring individual colonies to a 96-well steel target plate (Bruker Daltonics, Solna, Sweden) and adding 1 µL 70% formic acid on top of each sample. To the dried samples, 1 µL of 10 mg/mL α-cyano 4-hydroxycinnamic acid (HCCA) in matrix solution (Sigma-Aldrich) were added, and after evaporation the mass spectrum of each colony was determined by a matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) Microflex Instrument (Bruker Daltonics). The mass spectra were analyzed using the FlexControl and MALDI Biotyper 3.1 software with MBT Compass library, DB-6903 MSP (Bruker Daltonics) and compared to a combined library consisting of the MBT Compass library and an in-house reference database for the different hbs–LAB (HBS–LAB database) (Butler et al., Citation2016; Carbonnelle et al., Citation2011; Olofsson et al., Citation2014a). A score of 2.0 or greater was needed for positive hbs–LAB identification. All of the 13 initial hbs–LAB were represented among the analysed colonies. However, L. kunkeei Fhon2N was very clearly the dominant bacterium post-incubation, even though the incubation started with similar amounts of the 13 bacterial species.
Cell-free supernatant
The cell-free supernatant (CFS) from hbs–LAB mixture, as well as from uninoculated sMRS medium, was prepared as previously described (Butler et al., Citation2013), with minor modifications. The hbs–LAB mixture/sMRS media was centrifuged (4000 rpm, 30 min at 4 °C) and filter-sterilized (0.22 µm filter, Sarstedt, Nümbrecht, Germany) to get the CFS. After the filtration, 3 mL of CFS and sMRS were freeze-dried (Martin Christ Freeze Dryer, Osterode am Harz, Germany) overnight. The freeze-dried samples were kept at –80 °C and re-suspended in sterile water (300 µL) prior to use.
Agar well diffusion assay
Agar well diffusion assays (Alippi & Reynaldi, Citation2006; Butler et al., Citation2016) were used for initial assessment of the inhibitory activity of the hbs–LAB CFS on P. larvae. Soft MYPGP agar (0.8%) (15 mL) at 42 °C was inoculated with 150 µL of P. larvae bacterial suspension at McFarland standard 0.5 (OD600=0.08–0.10) and poured into a Petri dish to solidify. The wells of 5 mm diameter were cut into the agar and filled with 20 µL of the CFS. The plates were incubated at 35 °C with 5% CO2 for 72 h. The antimicrobial activity was quantified by measuring the radius of inhibition zones of each well.
Turbidimetric assay
Turbidimetric assays (Kavanagh, Citation1968) were used to assess the inhibitory activity of the hbs–LAB CFS on the germination and growth of P. larvae in liquid medium. For each assay, 5 mL MYPGP broth was supplemented with either 250 µL reconstituted CFS or sMRS media and inoculated with either spores or vegetative cells of P. larvae ERIC-I or ERIC-II (OD600=0.02–0.03). Un-supplemented MYPGP broth was used as a blank. The starting pH of the different media was 7.11, 6.86 and 5.86 for MYPGP, MYPGP + sMRS and MYPGP + CFS, respectively, which is well within the optimum pH range for the germination of P. larvae spores (Alvarado, Phui, Elekonich, & Abel-Santos, Citation2013). Bacterial growth was monitored by spectrophotometry at OD 600 nm (GYNESYS, ThermoSpectronic, USA) for 8 days. Experiments were repeated three times and the generation time of the bacteria was calculated as described previously (Monod, Citation1949).
Exposure bioassays
Larval exposure bioassays were used to assess the inhibitory activity of the hbs–LAB CFS on P. larvae pathogenesis in honey bee larvae. First instar worker larvae from honey bee colonies located at the Swedish University of Agricultural Sciences, Uppsala, Sweden were grafted with a Chinese grafting tool (Bienenzuchtgeräte, Graze, Weinstadt, Germany) into individual wells of 48-well tissue culture plates, each containing 10 µL of prewarmed diet (Aupinel et al., Citation2005). Each bioassay included technical controls for assessing the background larval mortality. Bioassays with >15% background mortality were excluded from the data. The experimental larvae were infected with 1 × 104 P. larvae spores per larvae: an infectious dose that produces sufficient larval mortality (about 70%) to observe the remedial effect of hbs–LAB treatment on larval mortality (Forsgren et al., Citation2010). Different experimental groups were fed larval diet supplemented with either the CFS (3 µL of reconstituted CFS in 10 µL diet) or a mixture of live 13 hbs–LAB cell suspension (1 × 107 bacteria per 10 µL diet) for 6 consecutive days, with increasing amounts of daily diet (10, 10, 20, 30, 40 and 60 µL food/larva/day, respectively). Control bioassays assessing any direct effect of live hbs–LAB on uninfected larvae were all negative. The larvae were maintained in an incubator at 35 °C with a relative humidity of 96%. The larvae were checked daily for mortality before feeding. Dead larvae were removed, the gut content streaked onto MYPGP agar for the cultivation of P. larvae with the identity of the bacterial colonies confirmed by PCR (Dobbelaere, de Graaf, Peeters, & Franciscus, Citation2001). Each bioassay was repeated three times for both P. larvae genotypes (ERIC-I and II), with a total of 30 larvae per replicate, except for the P. larvae-infected experimental group treated with live hbs–LAB bacteria, which was replicated twice (Supplementary Online Table 1).
Statistical analyses
All statistical analyses were performed using R (R Core Team, Citation2016). In agar well diffusion assays, the differences between inhibition zones of P. larvae genotypes were tested with an unpaired t-test. In the turbidimetric assay we were interested in the strength of the growth increase over time in control samples (only containing P. larvae) depending on the P. larvae genotypes, the growth stages (spores and vegetative cells), and their interactions. A subset containing all measurements were made until the observed maximum, plus two more time points. We used a linear mixed model (package lme4; Bates, Maechler, Bolker, & Walker, Citation2014) with the three replications as random effect and the three predictors as fixed effect and all interactions. To more appropriately model the increase the continuous variable time was fit as first order polynomial (after bootstrapping with the package ez (Lawrence, Citation2012) the estimates were back-transformed). We then compared the resulting slopes for each absorbance over times (package lsmeans (Lenth, Citation2016)). The exposure bioassays were analyzed with a generalized linear mixed model with a binomial likelihood (dead vs. alive; logit link), the treatment, the genotypes, and their interaction as explanatory variables, and the three replicates as random effect (meaning each dead vs. alive pair received its own likelihood). Here we compared the means (package multcomp Hothorn, Bretz, & Westfall, Citation2008; single step adjustment of P values).
Results
The purpose of these experiments was to determine whether the inhibitory effect of hbs–LAB on P. larvae seen previously (Forsgren et al., Citation2010) was due to secreted compounds produced by the hbs–LAB community, and to assess which aspects of P. larvae growth were affected. A cell-free supernatant (CFS) derived from the communal cultivation of 13 hbs–LAB, the secretome, was therefore tested for inhibitory activity against P. larvae in several different assays.
In vitro inhibition of P. larvae genotypes
The agar well diffusion assays showed a strong inhibition by the CFS on the growth of both P. larvae genotypes, resulting in clear and sharp inhibition zones. The ERIC-I genotype was considerably more inhibited than ERIC-II genotype (mean ± SE: ERIC-I: 4.00 mm ± 0.17, ERIC-II: 3.13 mm ± 0.17; t test: P = 0.02).
To assess the effect of the CFS on the P. larvae spore germination, vegetative growth and sporulation in liquid medium we used a turbidimetric bacterial growth assay. The method was run for both P. larvae genotypes, initiated either with endospores or vegetative cells (). Following spore germination, the P. larvae growth curve was characterized by a rapid vegetative growth, with estimated generation times of ∼48.4 and 18.7 min for the ERIC-I and ERIC-II genotypes, respectively. A sharp peak was observed followed by a decline in cell density as the bacterial culture entered the sporulation phase. However, in CFS-supplemented media, this vegetative growth peak was entirely absent, with the P. larvae growth curve increasing only slightly and very slowly over the same time period, reaching far lower final cell densities than in the non-supplemented media (; ). In spore-initiated cultures, the growth peak was delayed by ∼32 h (ERIC-I) and ∼24 h (ERIC-II) relative to vegetative cell-initiated cultures (; in red), corresponding to the time the spores needed to germinate before the vegetative cell growth starts. The ERIC-II P. larvae genotype also germinated considerably faster than the ERIC-I genotype. Also the sporulation phase was different for the two genotypes, with the ERIC-I genotype entering a much steeper and extensive decline than the ERIC-II genotype. The four CFS-supplemented growth curves were similar in shape () and very flat, indicating extensive inhibition of P. larvae growth. Since spore-initiated and vegetative cell-initiated cultures were equally inhibited, the inference was that the CFS primarily inhibited vegetative growth, rather than spore germination. Due to the absence of significant vegetative growth in CFS supplemented media, no inference could be made on the possible inhibition of sporulation by the CFS.
Figure 1. Inhibitory effect of hbs–LAB CFS on P. larvae growth in broth. Growth curves (black symbols and black lines) of P. larvae in MYPGP broth supplemented with hbs–LAB cell-free supernatant (P. larvae+CFS) compared to the controls (P. larvae). Bacterial growth was reflected by increasing turbidity, and measured by the optical density at 600 nm (OD600). The growth curves were initiated with either P. larvae ERIC-I spores (A), P. larvae ERIC-I vegetative cells (B), P. larvae ERIC-II spores (C), or P. larvae ERIC-II vegetative cells (D). In red: The strength of the growth increase of P. larvae was compared among all four combinations. Circles show the original subset and lines indicate model predictions with bootstrapped confidence limits. Different upper-case letters indicate significantly different slopes (Tukey contrast; p < 0.05).
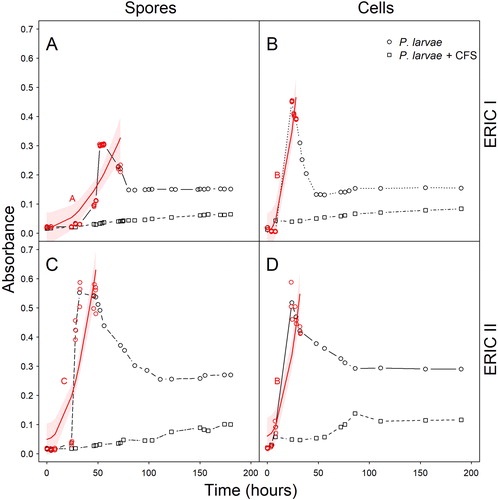
Table 1. Analysis-of-deviance tables (type III test) from models investigating the effect on absorbance in the turbidimetric assay (linear mixed model, LMM) and the survival of larvae (generalized linear mixed model, GLMM).
In vivo inhibition of P. larvae genotypes
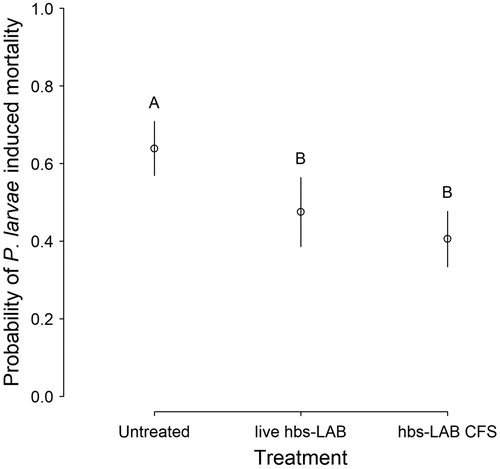
In vivo larval exposure bioassays revealed that the effect of both the live hbs–LAB and CFS treatments on reducing P. larvae-caused larval mortality were significant (; ; Supplementary Online Table 2). The direction and extent of the effect is consistent for both genotypes and similar to that recorded previously (Forsgren et al., Citation2010). Furthermore, the CFS treatment may have a superior inhibitory effect than live hbs–LAB treatment on larval mortality caused by P. larvae infection, especially for ERIC I genotypes (Supplementary Online Figure 1; Supplementary Online Table 3).
Figure 2. Inhibitory effect of hbs–LAB cell-free supernatant on P. larvae in larval bioassays. Probability of mortality for P. larvae-infected honey bee larvae treated with hbs–LAB cell-free supernatant (CFS), relative to untreated (negative control) and live hbs–LAB-treated (positive control) P. larvae-infected larvae. Symbols show predicted marginal means with CIs and different upper case letters indicate significant differences among the treatments (Tukey contrast; p < 0.01).
Discussion
Although much research has been conducted on various aspects of AFB and its causative agent, P. larvae, there is little information about the basic germination and growth characteristics of P. larvae, or how these are affected by putative antimicrobial agents. These are important parameters for the management of AFB, since the speed of spore germination and rate of P. larvae vegetative growth influences how fast the larva will die (Genersch, Ashiralieva, & Fries Citation2005; Yue, Nordhoff, Wieler, & Genersch Citation2008). If this rapid death occurs prior to capping, it will facilitate the detection and removal of the larval corpse by bee hygienic behavior. Delayed germination or vegetative growth enhances the probability that the diseased larva is capped, making hygienic detection and removal less likely and allowing sporulation to be completed. Any successful AFB treatment or management strategy has to take into account how it affects the triggers and timing of the major bacterial life-cycle events in relation to larval development and bee hygienic behavior.
The main result of our experiments is that the inhibitory effect of hbs–LAB on the growth and larval mortality caused by P. larvae appears to reside largely in extracellular, secreted substances (the “secretome"), and that these primarily (and strongly) inhibit the vegetative growth of P. larvae. This reduces bacterial proliferation and perhaps avoids the triggers for sporulation, and this is probably the primary reason for the enhanced survival of P. larvae-infected larvae treated with hbs–LAB CFS. There was no evidence that the CFS specifically inhibited or delayed spore germination, since the inhibitory effects were identical for spore and vegetative cell initiated turbidimetric cultures. However, it would also be incorrect to conclude that the CFS cannot affect spore germination: other types of experiments are required to verify or exclude this.
There are a number of metabolites and peptides secreted by hbs–LAB that could be responsible for the inhibitory effect. hbs–LAB secret a wide variety of antimicrobial substances and each species releases its own unique set of metabolites (Butler et al., Citation2013; Olofsson et al., Citation2014b). A common feature of all hbs–LAB is that they produce organic acids such as lactic, acetic and formic acid (Olofsson et al., Citation2014b). Previous studies have demonstrated that organic acids produced by Lactobacillus plantarum, Lactobacillus brevis (Mudroňová et al., Citation2011) and Lactobacillus johnsonii (Audisio et al., Citation2011) inhibit P. larvae vegetative growth. It has also been shown that the spores of Bacillus cereus were more resistant to organic acids than the vegetative cells, with the resistance furthermore also affected by pH (Wong & Chen, Citation1988). It is also highly likely that the hbs–LAB supernatant contains antimicrobial substances other than organic acids that contribute to the inhibitory effect on P. larvae. For example, hbs–LAB subjected to molecules associated with pathogen-associated microbial patterns (PAMPs), such as lipopolysaccharide, lipoteichoic acid and peptidoglycans produce a distinct and unique range of extracellular proteins and enzymes with possible antimicrobial activity. For example, L. helsinborgensis bacteriocin, a putative helveticin J homolog, was recently discovered (Butler et al., Citation2013). It is possible that some of the antimicrobial activity of honey (Butler et al., Citation2013; Olofsson et al., Citation2014b; Olofsson & Vásquez, Citation2008), including against P. larvae (Erler, Denner, Bobiş, Forsgren, & Moritz, Citation2014) can be traced to metabolites and peptides produced by hbs–LAB and that this may be part of the honey bee’s natural innate immunity against pathogens (Erler et al., Citation2014). The results presented here are similar to other studies demonstrating delayed spore germination or growth inhibition of pathogenic bacteria (e.g. Bacillus cereus, Wong and Chen, Citation1988; Bacillus anthracis, Gut, Prouty, Ballard, van der Donk, & Blanke, Citation2008) by certain specific LAB (e.g. Streptococcus lactis, Streptococcus thermophilus, Lactobacillus acidophilus, Lactobacillus bulgaricus) or their antimicrobial products (e.g. nisin; an antimicrobial peptide produced by Lactococcus lactis, Gut et al., Citation2008). The next stage in this research would be to identify and isolate the active substances produced and secreted by the hbs–LAB involved in the inhibition of P. larvae.
A second point of interest is the differential response of the two main P. larvae genotypes during our experiments. These genotypes are well characterized at both the biological and molecular level (Genersch et al., Citation2005; Heyndrickx & Vos, Citation1996; Poppinga & Genersch, Citation2015) and the results of these experiments in by-and-large confirm the biological characteristics of these genotypes. For example, ERIC-I genotypes are associated with spore production and elevated virulence at the colony level, while ERIC-II genotypes are associated with vegetative growth and elevated virulence at the individual level (Genersch et al., Citation2005; Poppinga & Genersch, Citation2015). This is consistent with the observations made here of a faster germination, shorter generation time, higher final cell density and delayed/reduced sporulation for ERIC-II (i.e. dominant vegetative stage) compared to ERIC-I (i.e. dominant spore stage). To this can be added that the CFS appears to be slightly more effective against the ERIC-I genotype than the ERIC-II genotype in the agar well diffusion assay, the larval exposure bioassay and the turbidimetric assay. This suggests that the vegetative state of the ERIC-II genotype maybe more robust to the natural antimicrobial agents produced by the hbs–LAB than the vegetative state of the ERIC-I genotype, which in turn may explain why ERIC-II has a more dominant vegetative state both in vitro and in vivo (Yue et al., Citation2008).
Conclusion
Considering the negative effects of antibiotics on commensal microbiota, the evolution of resistance of P. larvae to conventional antibiotic treatment (Alippi et al., Citation2007; Evans, Citation2003; Miyagi et al., Citation2000), and the overall risk from antibiotic use for both animals and humans (Al-Waili et al., Citation2012), our research findings are an important first step for using beneficial bacteria against AFB in honey bee colonies which needs to be further studied in the future.
Supplemental Material
Download JPEG Image (187 KB)Supplemental Tables and Figure caption
Download MS Word (18.8 KB)Acknowledgements
The authors also wish to thank Emilia Semberg and Anette Mårtensson for laboratory assistance.
Additional information
Funding
References
- Al-Waili, N., Salom, K., Al-Ghamdi, A., & Ansari, M. J. (2012). Antibiotic, pesticide, and microbial contaminants of honey: Human health hazards. Scientific World Journal, 2012, 1–9. doi: 10.1100/2012/930849
- Alippi, A. M., López, A. C., Reynaldi, F. J., Grasso, D. H., & Aguilar, O. M. (2007). Evidence for plasmid-mediated tetracycline resistance in Paenibacillus larvae, the causal agent of American Foulbrood (AFB) disease in honey bees. Veterinary Microbiology, 125, 290–303. doi: 10.1016/j.vetmic.2007.05.018
- Alippi, A. M., & Reynaldi, F. J. (2006). Inhibition of the growth of Paenibacillus larvae, the causal agent of American foulbrood of honey bees, by selected strains of aerobic spore-forming bacteria isolated from apiarian sources. Journal of Invertebrate Pathology, 91, 141–146. doi: 10.1016/j.jip.2005.12.002
- Alvarado, I., Phui, A., Elekonich, M. M., & Abel-Santos, E. (2013). Requirements for in vitro germination of Paenibacillus larvae spores. Journal of Bacteriology, 195, 1005–1011. doi: 10.1128/JB.01958-12
- Animal and Plant Health Agency. (2017). Foulbrood disease of honey bees and other common brood disorders, UK.
- Audisio, M. C., Torres, M. J., Sabaté, D. C., Ibarguren, C., & Apella, M. C. (2011). Properties of different lactic acid bacteria isolated from Apis mellifera L. bee-gut. Microbiological Research, 166, 1–13. doi: 10.1016/j.micres.2010.01.003
- Aupinel, P., Fortini, D., Dufour, H., Taséi, J. N., Michaud, B., Odoux, J. F., & Pham-Delègue, M. H. (2005). Improvement of artificial feeding in a standard in vitro method for rearing Apis mellifera larvae. Bulletin of Insectology, 58, 107–111.
- Bamrick, J. (1967). Resistance to American Foulbrood in honey bees VI. Spore germination in larvae of different ages. Journal of Invertebrate Pathology, 9, 30–34. doi: 10.1016/0022-2011(67)90039-0
- Bates, D. M., Maechler, M., Bolker, B. M., & Walker, S. (2014). lme4: Linear mixed-effects models using Eigen and S4. R Package Version 1.1-7. http://CRAN.R-project.org/package=lme4.
- Benitez, L. B., Velho, R. V., de Souza da Motta, A., Segalin, J., & Brandelli, A. (2012). Antimicrobial factor from Bacillus amyloliquefaciens inhibits Paenibacillus larvae, the causative agent of American foulbrood. Archives of Microbiology, 194, 177–185. doi: 10.1007/s00203-011-0743-4
- Butler, È., Alsterfjord, M., Olofsson, T. C., Karlsson, C., Malmström, J., & Vásquez, A. (2013). Proteins of novel lactic acid bacteria from Apis mellifera mellifera: An insight into the production of known extra-cellular proteins during microbial stress. BMC Microbiology, 13, 235–246. doi: 10.1186/1471-2180-13-235
- Butler, È., Oien, R. F., Lindholm, C., Olofsson, T. C., Nilson, B., & Vásquez, A. (2016). A pilot study investigating lactic acid bacterial symbionts from the honeybee in inhibiting human chronic wound pathogens. International Wound Journal, 13, 729–737. doi: 10.1111/iwj.12360
- Carbonnelle, E., Mesquita, C., Bille, E., Day, N., Dauphin, B., Beretti, J. L., Ferroni, A.,… Nassif, X. (2011). MALDI–TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clinical Biochemistry, 44, 104–109. doi: 10.1016/j.clinbiochem.2010.06.017
- Cogliani, C., Goossens, H., & Greko, C. (2011). Restricting antimicrobial use in food animals: Lessons from Europe. Microbe Journal, 6, 274–279. doi: 10.1128/microbe.6.274.1
- Dobbelaere, W., de Graaf, D., Peeters, J., & Franciscus, J. (2001). Development of a fast and reliable diagnostic method for American foulbrood disease (Paenibacillus larvae subsp. larvae) using a 16S rRNA gene based PCR. Apidologie, 32, 363–370. doi: 10.1051/apido:2001136
- Elzen, P. J., Westervelt, D., Causey, D., Ellis, J., Hepburn, H. R., & Neumann, P. (2002). Method of application of tylosin, an antibiotic for American foulbrood control, with effects on small hive beetle (Coleoptera: Nitidulidae) populations. Journal of Economic Entomology, 95, 1119–1122. doi: 10.1603/0022-0493-95.6.1119
- Erler, S., Denner, A., Bobiş, O., Forsgren, E., Moritz, R. F. A. (2014). Diversity of honey stores and their impact on pathogenic bacteria of the honeybee, Apis mellifera. Ecology & Evolution, 4, 3960–3967. doi: 10.1002/ece3.1252
- Evans, J. D. (2003). Diverse origins of tetracycline resistance in the honeybee bacterial pathogen Paenibacillus larvae. Journal of Invertebrate Pathology, 83, 46–50. doi: 10.1016/S0022-2011(03)00039-9
- Evans, J. D., & Lopez, D. L. (2004). Bacterial probiotics induce an immune response in the honeybee (Hymenoptera: Apidae). Journal of Economic Entomology, 97, 752–756. doi: 10.1093/jee/97.3.752
- Forsgren, E., Olofsson, T. C., Vásquez, A., & Fries, I. (2010). Novel lactic acid bacteria inhibiting Paenibacillus larvae in honeybee larvae. Apidologie, 41, 99–108. doi: 10.1051/apido/2009065
- Forsgren, E., Stevanovic, J., Fries, I. (2008). Variability in germination and in temperature and storage resistance among Paenibacillus larvae genotypes. Veterinary Microbiology, 129, 342–349. doi: 10.1016/j.vetmic.2007.12.001
- Fries, I., Lindström, A., & Korpela, S. (2006). Vertical transmission of American foulbrood (Paenibacillus larvae) in honey bees (Apis mellifera). Veterinary Microbiology, 114, 269–274. doi: 10.1016/j.vetmic.2005.11.068
- Gallai, N., Salles, J. M., Settele, J., & Vaissière, B. E. (2009). Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecological Economics, 68, 810–821. doi: 10.1016/j.ecolecon.2008.06.014
- Genersch, E. (2010a). American Foulbrood in honey bees and its causative agent, Paenibacillus larvae. Journal of Invertebrate Pathology, 103, S10–S19. doi: 10.1016/j.jip.2009.06.015
- Genersch, E. (2010b). Honey bee pathology: Current threats to honey bees and beekeeping. Applied Microbiology & Biotechnology, 87, 87–97. doi: 10.1007/s00253-010-2573-8
- Genersch, E., Ashiralieva, A., & Fries, I. (2005). Strain- and genotype-specific differences in virulence of Paenibacillus larvae subsp. larvae, a bacterial pathogen causing American foulbrood disease in honey bees. Applied & Environmental Microbiology, 71, 7551–7555. doi: 10.1128/AEM.71.11.7551-7555.2005
- Genersch, E., Forsgren, E., Pentikäinen, J., Ashiralieva, A., Rauch, S., Kilwinski, J., … Fries, I. (2006). Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without subspecies differentiation. International Journal of Systematic & Evolutionary Microbiology, 56, 501–511. doi: 10.1099/ijs.0.63928-0
- Genersch, E., & Otte, C. (2003). The use of repetitive element PCR fingerprinting (rep-PCR) for genetic subtyping of German field isolates of Paenibacillus larvae subsp. larvae. Apidologie, 34, 195–206. doi: 10.1051/apido:2003025
- Gut, I. M., Prouty, A. M., Ballard, J. D., van der Donk, W. A., & Blanke, S. R. (2008). Inhibition of Bacillus anthracis spore outgrowth by nisin. Antimicrobial Agents & Chemotherapy, 52, 4281–4288. doi: 10.1128/AAC.00625-08
- Heyndrickx, M., & Vos, P. D. E. (1996). Reclassification of Paenibacillus (formerly Bacillus) pulvifaciens (Nakamura 1984) Ash et al. 1994, a later subjective synonym of Paenibacillus (formerly Bacillus) larvae (White 1906) Ash et al. 1994, as a subspecies of P. larvae, with emended description. International Journal of Systematic & Evolutionary Microbiology, 46, 270–279. doi: 10.1099/00207713-46-1-270
- Hothorn, T., Bretz, F., & Westfall, P. (2008). Simultaneous inference in general parametric models. Biometrical Journal, 50, 346–363. doi: 10.1002/bimj.200810425
- Kavanagh, F. (1968). Turbidimetric assays: The antibiotic dose–response line. Applied Microbiology, 16, 777–780.
- Killer, J., Dubná, S., Sedláček, I., Švec, P., 2014. Lactobacillus apis sp. nov., from the stomach of honey bees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. International Journal of Systematic & Evolutionary Microbiology, 64, 152–157. doi: 10.1099/ijs.0.053033-0
- Lamei, S., Hu, Y. O. O., Olofsson, T. C., Andersson, A. F., Forsgren, E., & Vásquez, A. (2017). Improvement of identification methods for honeybee specific lactic acid bacteria: Future approaches. PLoS One, 12, e0174614. doi: 10.1371/journal.pone.0174614
- Lawrence, M. A. (2012). ez: Easy analysis and visualization of factorial experiments. R package version 4.1-1.
- Lenth, R. V. (2016). Least-squares means: The R package lsmeans. Journal of Statistical Software, 69, 1–33.
- Maggi, M., Negri, P., Plischuk, S., Szawarski, N., De Piano, F., De Feudis, L., … Audisio, C. (2013). Effects of the organic acids produced by a lactic acid bacterium in Apis mellifera colony development, Nosema ceranae control and fumagillin efficiency. Veterinary Microbiology, 167, 474–483. doi: 10.1016/j.vetmic.2013.07.030
- McNally, L., Viana, M., Brown, S. P. (2014). Cooperative secretions facilitate host range expansion in bacteria. Nature Communications, 5, 1–8.
- Minnaard, J., & Alippi, A. M. (2016). Partial characterization of bacteriocin-like compounds from two strains of Bacillus cereus with biological activity against Paenibacillus larvae, the causal agent of American Foulbrood disease. Applied Microbiology, 63, 442–449. doi: 10.1111/lam.12665
- Miyagi, T., Peng, C. Y., Chuang, R. Y., Mussen, E. C., Spivak, M. S., & Doi, R. H. (2000). Verification of oxytetracycline-resistant American foulbrood pathogen Paenibacillus larvae in the United States. Journal of Invertebrate Pathology, 75, 95–96. doi: 10.1006/jipa.1999.4888
- Monod, J. (1949). The growth of bacterial cultures. Annual Review of Microbiology, 3, 371–394. doi: 10.1146/annurev.mi.03.100149.002103
- Mudroňová, D., Toporčák, J., Nemcová, R., Gancarčíková, S., Hajdučková, V., & Rumanovská, K. (2011). Lactobacillus sp. as a potential probiotic for the prevention of Paenibacillus larvae infection in honey bees. Journal of Apicultural Research, 50, 323–324. doi: 10.3896/IBRA.1.50.4.11
- Nordström, S., & Fries, I. (1995). A comparison of media and cultural conditions for identification of Bacillus larvae in honey. Journal of Apicultural Research, 34, 97–103. doi: 10.1080/00218839.1995.11100894
- Olofsson, T. C., Alsterfjord, M., Nilson, B., Butler, È., & Vásquez, A. (2014a). Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. International Journal of Systematic & Evolutionary Microbiology, 64, 3109–3119. doi: 10.1099/ijs.0.059600-0
- Olofsson, T. C., Butler, E., Markowicz, P., Lindholm, C., Larsson, L., & Vásquez, A. (2014b). Lactic acid bacterial symbionts in honey bees—An unknown key to honeys antimicrobial and therapeutic activities. International Wound Journal, 13, 668–679. doi: 10.1111/iwj.12345
- Olofsson, T. C., & Vásquez, A. (2008). Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Current Microbiology, 57, 356–363. doi: 10.1007/s00284-008-9202-0
- Poppinga, L., & Genersch, E. (2015). Molecular pathogenesis of American Foulbrood: How Paenibacillus larvae kills honeybee larvae. Current Opinion in Insect Science, 10, 29–36. doi: 10.1016/j.cois.2015.04.013
- R Core Team (2016). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. http://www.R-project.org/.
- Sabaté, D. C., Carrillo, L., Audisio, M. C. (2009). Inhibition of Paenibacillus larvae and Ascosphaera apis by Bacillus subtilis isolated from honeybee gut and honey samples. Research in Microbiology, 160, 193–199. doi: 10.1016/j.resmic.2009.03.002
- Vásquez, A., Forsgren, E., Fries, I., Paxton, R. J., Flaberg, E., Szekely, L., & Olofsson, T. C. (2012). Symbionts as major modulators of insect health: Lactic acid bacteria and honey bees. PLoS One, 7, e33188. doi: 10.1371/journal.pone.0033188
- Wong, H., & Chen, Y. (1988). Effects of lactic acid bacteria and organic acids on growth and germination of Bacillus cereus. Applied & Environmental Microbiology, 54, 2179–2184.
- Woodrow, A. W. (1942). Susceptibility of honeybee larvae to individual inoculations with spores of Bacillus larvae. Journal of Economic Entomology, 35, 892–895. doi: 10.1093/jee/35.6.892
- Yoshiyama, M., & Kimura, K. (2009). Bacteria in the gut of Japanese honeybee, Apis cerana japonica, and their antagonistic effect against Paenibacillus larvae, the causal agent of American foulbrood. Journal of Invertebrate Pathology, 102, 91–96. doi: 10.1016/j.jip.2009.07.005
- Yue, D., Nordhoff, M., Wieler, L. H., & Genersch, E. (2008). Fluorescence in situ hybridization (FISH) analysis of the interactions between honeybee larvae and Paenibacillus larvae, the causative agent of American foulbrood of honey bees (Apis mellifera). Environmental Microbiology, 10, 1612–1620. doi: 10.1111/j.1462-2920.2008.01579.x
