ABSTRACT
Scaphander gracilis Watson, 1883 is a deep-sea Cephalaspidea gastropod species endemic to the Azores in the mid-North Atlantic, and which was up to now only known from empty shells with a unique thick callus. Here, we describe for the first time the morphology of this snail species from anatomical dissections and provide a DNA barcode. A phylogeny based on the gene marker cytochrome c oxidase subunit I was inferred to test the position of S. gracilis among the genus Scaphander. In addition, the ABGD species delimitation method was employed and uncorrected p-distances were estimated. The sister relationship of S. gracilis was not resolved, but it clustered with the Atlantic species S. meridionalis and S. nobilis and the Pacific species S. grandis, S. mundus, and S. subglobosus. The smallest genetic distance was to the NE Pacific species S. grandis (4.29% in COI). Foraminifera were found in the digestive tract and the species is confirmed alive at depths between 2065−2066 m. The fact that S. gracilis was found on both sides of the Mid-Atlantic Ridge suggests that this significant geological phenomenon is not an insurmountable barrier for dispersal in this species.
Introduction
The deep sea (waters and bottoms below 200 m depth) is the largest ecosystem on the planet and covers 65% of the Earth (Thistle Citation2003; Tyler Citation2003; Jobstvogt et al. Citation2014). It was long thought to host little or no life (Tyler Citation2003), but today we know that a high diversity is found in the deep oceans (Grassle and Maciolek Citation1992; Van Dover Citation2000). In fact, an increasing number of studies have investigated deep-sea biodiversity in several regions of the world, but often these studies focus on a reduced number of taxa and are typically characterised by a limited spatial scale of investigation (e.g.: regional species diversity off the eastern US coast (Etter and Grassle Citation1992; Grassle and Maciolek Citation1992); species diversity in the Mediterranean (Danovaro et al. Citation2010); Scaphander snails in the Atlantic (Eilertsen and Malaquias Citation2013a, Citation2015)), and overall deep-sea benthic diversity, particularly of abyssal plains, remains poorly known.
One deep-sea benthic faunal group that received considerable attention in recent years in the Atlantic Ocean was the gastropod genus Scaphander Montfort, Citation1810 (Eilertsen and Malaquias Citation2013a, Citation2013b, Citation2015; Siegwald et al., Citation2020). Scaphander includes 33 species worldwide (MolluscaBase Citation2021) that inhabit predominantly deep-sea soft bottoms where they feed preferentially upon foraminiferans, including agglutinating forms, but also bivalves, gastropods, scaphopods, and polychaetes with calcareous tubes (Eilertsen and Malaquias Citation2013b). Ten species are recognised in the Atlantic Ocean, seven of those have representative DNA barcodes. The species Scaphander clavus Dall, Citation1889 is only known from its shell and anatomy, Scaphander gracilis Watson, Citation1883 is only known from empty shells (Eilertsen and Malaquias Citation2013a; Siegwald et al. Citation2020) and S. imperceptus Bouchet, Citation1975 is only known from old material (Citation1883) not suitable for regular DNA extraction methods.
Scaphander gracilis is a rare species endemic to the Archipelago of the Azores in the mid-North Atlantic Ocean, and was described from empty shells that have a distinctive parietal callus. Shells have been collected between 1299 and 2995 m depth, but no complete specimen including soft tissue, has ever been collected (Watson Citation1883; Dautzenberg and Fischer Citation1896; Locard Citation1897; Bouchet Citation1975; Eilertsen and Malaquias Citation2013a). The current work aims to contribute to a better understanding of Atlantic deep-sea molluscan biodiversity by providing the first data on the anatomy and ecology of S. gracilis. An inference of relationships for this species within its genus using molecular phylogenetics is discussed as is the biogeography of this species.
Material and methods
One specimen of S. gracilis was collected and preserved in 96% ethanol during the Meteor cruise M150, Biodiversity of the Azores (BIODIAZ) in 2018, an expedition led by the Senckenberg am Meer, German Center for Marine Biodiversity Research. The specimen was dissected for examination of the digestive and reproductive systems, which were drawn using a stereo microscope fitted with a camera lucida. The buccal bulb was dissolved in a Proteinase K solution composed of 20 μL Proteinase K and 180 μL buffer ATL obtained from the Qiagen DNeasy Blood and Tissue kit, and incubated at 56°C overnight (protocol modified from Holznagel Citation1998; Vogler Citation2013). The gut content was collected under a stereo microscope then visualised with SEM. The male reproductive system was dissected and the penial chamber was critical-point dried for SEM analysis. The shell was photographed using a DSLR camera equipped with macro lens. Details of the shell and gizzard plates were photographed using a Leica M205C stereo microscope equipped with a Leica DMC5400 camera, and images were stacked using Zerene Stacker 1.04 software (http://zerenesystems.com/cms/home). The radula, penial chamber section and gut content were coated with gold-palladium and examined with a ZEISS SUPRA 55VP electron scanning microscope (SEM) at the Department of Earth Science (University of Bergen).
DNA was extracted from foot tissue using the Qiagen DNeasy Blood and Tissue Kit following the manufacturer’s instructions. The barcoding gene marker cytochrome c oxidase subunit I (COI) was amplified and purified following the methods described by Eilertsen and Malaquias (Citation2013a). Purified products were sequenced using an ABI 3730XL DNA Analyser (Applied Biosystems). The software programme Geneious (v. Prime 2021.1.1) was used to inspect, edit and assemble the chromatograms. The sequences were blasted in GenBank to check for potential contamination. An additional 23 COI sequences were pulled from GenBank (22 Scaphandridae and one Bulla vernicosa for outgroup). The final dataset () was edited and aligned using MUSCLE (Edgar Citation2004) implemented in Geneious and the quality of the alignment was assessed by translating to proteins and checking for stop-codons. Saturation of codon positions was assessed by plotting uncorrected p-distances against total substitutions (transversions + transitions) using MEGA 10.0.1 (Kumar et al. Citation2018). The best-fitting model of evolution was selected using Akaike’s information criterion (Akaike Citation1974) implemented in jModelTest 2.1.10 (Darriba et al. Citation2012). A Bayesian Inference analysis was performed in MrBayes 3.2.6 (Huelsenbeck and Ronquist Citation2001) with three parallel runs of five million generations and sampling every 100 generations. Convergence was assessed in Tracer 1.7.1 (Rambaut et al. Citation2018) for each run by plotting the likelihood against the number of generations, with a burn-in set to 10%. Tree annotation was done in FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree), and adjustments were made in Adobe Illustrator CS6.
Table 1. Taxon set used for molecular phylogenetics with locality, voucher numbers, and GenBank accession numbers (* novel sequences generated for this study)
The molecular species delimitation method Automatic Barcode Gap Discovery (ABGD; Puillandre et al. Citation2012) was used based on the alignment. The web version of the ABGD programme (available at https://bioinfo.mnhn.fr/abi/public/abgd/) was run for the three different models available (Jukes Cantor, Kimura, and Simple Distance) using default settings (Pmin = 0.001, Pmax = 0.1, 10 steps, 20 Nb bins) and the relative gap width (X) adjusted to 1.0.
Geographical distributions of known S. gracilis specimens were collected from the study of museum specimens, literature records that could confidently be attributed to the species, and newly collected material, and were plotted using R (R Core Team Citation2021). Shoreline data were taken from the Global Self-consistent, Hierarchical, High-resolution Geography Database (GSHHG v.2.3.7 https://www.soest.hawaii.edu/pwessel/gshhg; Wessel and Smith Citation1996). Mid-Atlantic Ridge data were taken from Bird (Citation2003).
Abbreviations
DBUA: Department of Biology of the University of the Azores, Ponta Delgada, Azores, Portugal; MCZ: Museum of Comparative Zoology, Harvard University, Boston, MA, USA; MIMB: Museum of the National Scientific Center of Marine Biology, Vladivostok, Russia; MNHN: Muséum national d’Histoire naturelle, Paris, France; MZUSP, Museu de Zoologia da Universidade de São Paulo, Brazil; NHMUK: The Natural History Museum, London, UK; NMINH: National Museum of Ireland (Natural History Division), Dublin, Ireland; RMNH: National Museum of Natural History (Naturalis), Leiden, The Netherlands; USNM: National Museum of Natural History [United States National Museum], Smithsonian Institution, Washington DC, USA; ZMBN: Department of Natural History, University Museum of Bergen, Norway; H: shell height, PP: posterior probability.
Results
Taxonomy
Order Cephalaspidea P. Fischer, Citation1883
Family Scaphandridae G.O. Sars, Citation1878
Genus Scaphander de Montfort, Citation1810
Scaphander gracilis Watson, Citation1883
(, )
Table 2. COI uncorrected p-distances between and within Scaphander species
Figure 1. Scaphander gracilis Watson, Citation1883. (a) apertural (left), adpertural (right), apical (bottom) views of shell from south off Flores Island, Azores, lectotype, NHMUK 1887.2.9.2183–6, H = 13.5 mm (images courtesy of the NHMUK photographic unit). (b) apertural (left), adpertural (right), apical (bottom) views of shell from south of São Miguel Island, Azores, paralectotype, NHMUK 1887.2.9.2187 − 8, H = 13.0 mm (images courtesy of the NHMUK photographic unit). (c) apertural (left), adpertural (right), apical (bottom) views of shell from between São Miguel and Santa Maria Islands, Azores, DBUA 1630, H = 20 mm. (d) stereo microscope image of the sculpture of the shell illustrated in C. Scale bar = 1 mm.
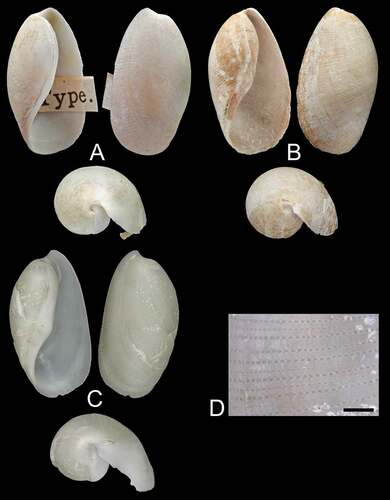
Figure 2. Anatomical details of Scaphander gracilis Watson, Citation1883 (DBUA 1630, H = 20 mm). (a) radula; (b) rachidian tooth; (c) denticulation of the inner edge of lateral teeth; (d) anterior part of digestive tract; (e) gizzard plates, inner side of the paired plates and lateral view of the unpaired plate; (f) male reproductive system; (g) lining of the penial chamber. bb, buccal bulb; c, crop; go, genital opening; m, mouth; o, oesophagus; p, prostate; pc, penial chamber; pd, prostatic duct; pgp, paired gizzard plates; sg, salivary gland; ugp, unpaired gizzard plate. Scale bars: A, G = 200 µm; B, C = 20 µm; D, F = 1 mm; E = 2 mm.
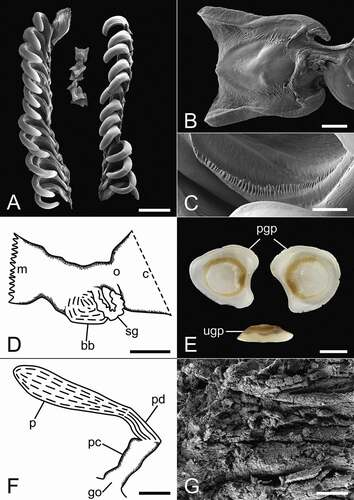
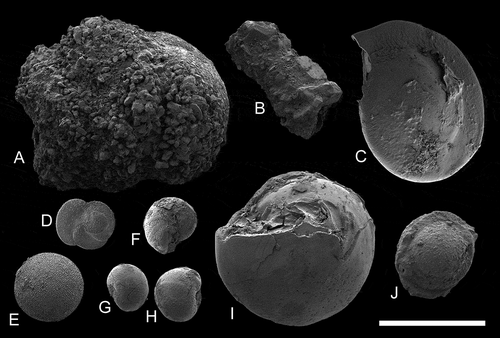
Figure 3. Scanning electron micrographs of foraminifera taken from the gut content of Scaphander gracilis (DBUA 1630, H = 20 mm). (a–b) agglutinating foraminifera; (c−j) calcareous foraminifera. Scale bar = 1 mm.
Scaphander gracilis Watson, Citation1883: 345−346; Watson, Citation1886: 645−646, pl. 47, –c; Pilsbry, Citation1893: 247−248, pl. 31, figs 19−20; Dautzenberg & Fischer, Citation1896: 402; Locard, Citation1897: 47–49, pl. 1, figs 15–18; Bouchet, Citation1975: 338, pl. 3, fig. i; Eilertsen & Malaquias, Citation2013a: 404−405, figs 2.10; 14.
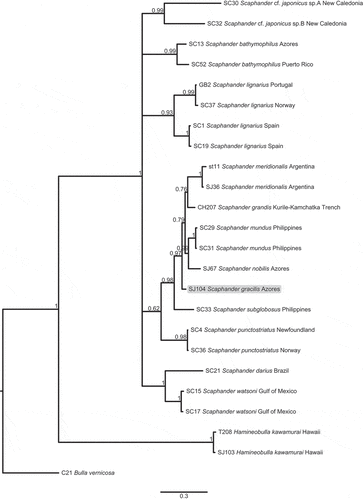
Figure 4. Bayesian phylogenetic tree based on partial sequences of the COI gene. Figures on nodes are posterior probabilities, scale bar refers to branch lengths. The tree was rooted using the species Bulla vernicosa.
Scaphander gracilis var. minor Locard, Citation1897: 47–49.
Scaphander gracilis var. major Locard, Citation1897: 47–49.
Type material
Western Azores, south of Flores Island (38°30ʹN, 31°14ʹW; 1829 m), 4 syntypes (images seen; https://data.nhm.ac.uk/object/80cd6292-170e-4636-9c32-859ad933b36e/1631750400000), lectotype and paralectotypes here designated (lectotype: H = 13.5 mm; paralectotypes: H = 8.8, 5.1, 11.3 mm), NHMUK 1887.2.9.2183–2186; Azores, south of São Miguel Island (37°26ʹN, 25°13ʹW; 1829 m), 2 syntypes (images seen; Natural History Museum Citation2014), specimens designated here as paralectotypes, NHMUK 1887.2.9.2187–2188 (H = 13.0, 14.4 mm) (Eilertsen and Malaquias Citation2013a); 1 syntype (image seen), here designated as paralectotype, NMINH 1899.114.182 (H = 14 mm).
Other material examined
Azores, between Santa Maria and São Miguel Islands (37°10.820ʹN, 025°11.750ʹW − 37°10.368ʹN, 025°12.957ʹW; 2065 − 2066 m), 1 spc. dissected and sequenced for the mitochondrial gene COI, DBUA 1630, H = 20 mm.
Revised diagnosis
Shell solid, white, pyriform. Parietal callus thick, white, smooth, can present an angular hump around aperture inflexion point. Spire concealed. Outer lip protruding posteriorly beyond apex. Spiral sculpture composed of medium and small punctuated striations. Prostate cylindrical, prostatic duct short, narrower than prostate. Penial chamber lined with soft longitudinal grooves. Penial papilla absent.
Shell ()
Maximum H = 24 mm (Locard Citation1897). Shell solid, white, pyriform, covered by thin white to pale yellow periostracum. Parietal callus thick, white, smooth, can present an angular hump around aperture inflexion point. Spire concealed. Outer lip protruding posteriorly beyond apex in small wing. Spiral sculpture composed of regular lines of small or medium-sized punctuations.
Radula ()−(c))
Radular formula 15 × 1.1.1. Lateral teeth hook-shaped, curved, with weak denticulation on inner edge. Small rachidian teeth tetragonal, with expanded outward corners, straight or curved.
Digestive system ()−(e))
Salivary glands long with uneven surface. Paired gizzard plates subtriangular to kidney-shaped; unpaired gizzard plate thinner, elongated.
Reproductive system ()−(g))
Prostate elongate, cylindrical, narrowing towards penial chamber. Prostatic duct short, narrower than prostate, separating prostate from penial chamber. Penial chamber cylindrical, lined with soft longitudinal ridges running from genital opening to prostatic duct. Penial papilla absent.
Ecology
Feeds on foraminifera, agglutinating and non-agglutinating, ingested along with sand and mud (). Live specimen collected between 2065 − 2066 m (present study), empty shells records between 1299 − 2995 m (Watson Citation1883; Dautzenberg and Fischer Citation1896; Locard Citation1897; Bouchet Citation1975; Eilertsen and Malaquias Citation2013a).
Distribution
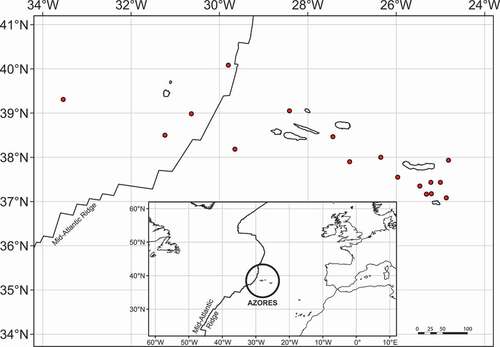
Only known from the Archipelago of the Azores, where it is reported west and southwards of Flores Island, on the western slope of the mid-Atlantic ridge (Watson Citation1883) and eastwards of the Islands of São Miguel and Santa Maria, on the eastern side of the mid-Atlantic ridge (Watson Citation1883; Dautzenberg and Fischer Citation1896; Locard Citation1897; Bouchet Citation1975; present study).
Molecular analysis and species delimitation
The COI alignment dataset included 658 bp, and the best-fitting model of evolution was TIM3 + I + G. The resulting COI tree clustered all Scaphander specimens together with maximum support (PP = 1). The S. gracilis specimen was part of a larger clade together with other Atlantic (S. meridionalis, S. nobilis) and Pacific species (S. grandis, S. mundus, S. subglobosus) (PP = 0.98; ). Sister relationships were generally poorly resolved and this was also the case for S. gracilis ().
COI uncorrected p-distances ranged from 0.18% to 1.25% within Scaphander species and from 4.29% to 20.04% between species. Genetic distances between S. gracilis and other Scaphander species varied from 4.29% to 19.14%. The smallest genetic distance was to the NE Pacific species S. grandis (4.29%), followed by the western Pacific species S. mundus (4.47%) and to the amphi-Atlantic species S. nobilis (6.62%).
The three independent models of evolution implemented in the ABGD method recovered 15 Scaphander lineages, including S. gracilis as a candidate species.
Discussion
Watson (Citation1883) described Scaphander gracilis based on shells found during the ‘Challenger’ expedition around the Azores Islands. He remarked on its similarities to the shells of S. punctostriatus (Mighels & C. B. Adams, Citation1842), but stressed the uniqueness of the striations and thick callus differentiating S. gracilis. Additional shells were subsequently found during various cruises around the Azores (e.g. ‘Hirondelle’ reported by Dautzenberg and Fischer (Citation1896); ‘Travailleur’ and ‘Talisman’ reported by Locard (Citation1897); ‘Charcot-Biaçores’ reported by Bouchet (Citation1975)).
Locard (Citation1897) described two varieties of S. gracilis: major, a larger and more pyriform variety; and minor, a smaller shell with a thicker callus, but Eilertsen and Malaquias (Citation2013a) attributed these differences to intraspecific variability. The shell of S. gracilis has been likened to the elongate form of the Atlantic Scaphander punctostriatus (Locard Citation1897; Marcus Citation1974), however, it is easily distinguishable from all other known Scaphander species by its thick callus, which even in the form without an angular hump, is conspicuously more pronounced than in other pyriform species of Scaphander.
Marcus (Citation1974) mentioned the occurrence of S. gracilis in the Caribbean, while at the same time expressed uncertainty about the identification of the specimens, stating that they could be S. punctostriatus, a known amphi-Atlantic species commonly found in the Caribbean. Eilertsen and Malaquias (Citation2013a) did not find evidence for the presence of the species in the western Atlantic and considered S. gracilis to be restricted to the Azores. This is also our opinion after comparison of shells between Caribbean and Azorean specimens.
Eilertsen and Malaquias (Citation2013a) recognised eight valid species of Scaphander in the Atlantic, and Chaban et al. (Citation2019) reassigned Meloscaphander imperceptus Bouchet, Citation1975 to Scaphander, to which Siegwald et al. (Citation2020) added S. meridionalis from offshore Argentina. Among these 10 species, S. gracilis was the only one known solely from shells. The present study describes for the first time the anatomy of S. gracilis and includes the first molecular sequence data from it; contributing to a preliminarily test of its phylogenetic position among Scaphander. Our illustration of the radula ()) may lead to the false impression that only few rows possess a rachidian tooth, but this is only because they are fragile and fall off easily during the cleaning and preparation for SEM. Another noteworthy feature is the comparatively larger size of the salivary glands when compared with other Atlantic species. Only S. bathymophilus and S. nobilis have similar or larger size glands (); see Eilertsen and Malaquias Citation2013a).
The COI uncorrected p-distances and the phylogenetic tree suggest a closer relationship to the other white sub-rectangular shelled Scaphander species, namely S. grandis, S. mundus, S. nobilis and S. meridionalis (; ), but the thickened smooth and often angular callus of S. gracilis makes it easy to distinguish from its congeners.
Interestingly, the known and restricted distribution of S. gracilis, which is only confirmed from deep waters in the Archipelago of the Azores, makes it the only Scaphander species with a geography limited to offshore Islands. The fact that S. gracilis was found on both sides of the Mid-Atlantic Ridge () suggests that this significant geological phenomenon is not an insurmountable barrier for dispersal in this species. The role of the Mid-Atlantic Ridge as a putative barrier for dispersal of bathyal and abyssal organisms across the western and eastern sides of the Atlantic realm remains poorly understood and heavily hampered by lack of molecular studies (Mullineaux et al. Citation2002; Zardus et al. Citation2006; Etter et al. Citation2011), and this work contributes to a greater understanding of the bathyal malacofauna of the Atlantic.
Acknowledgements
We are grateful to S. Ávila and R. Cordeiro (University of the Azores) for making specimens available for study, and to A. Salvador and K. Webb (NHMUK), as well as to A. Geraghty and P. Viscardi (NMINH) for providing type specimen images. We also thank I. Heggstad (Laboratory for Electron Microscopy, Department of Earth Science, University of Bergen) for help with critical-point drying and SEM work and L. Lindblom (DNA Lab, Department of Natural History, University of Bergen) for help with molecular sequencing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Akaike H. (1974). A new look at the statistical model identification. IEEE Trans. Automat. Contr., 19(6), 716–723. https://doi.org/10.1109/TAC.1974.1100705
- Bird P. 2003. An updated digital model of plate boundaries. Geochem Geophys Geosystems. 4(3):1027. doi:https://doi.org/10.1029/2001GC000252.
- Bouchet P. 1975. Opisthobranches de profondeur de l’océan Atlantique: i Cephalaspidea. Cah Biol Mar. 16(3):317–365.
- Chaban EM, Ekimova IA, Schepetov DM and Chernyshev AV. (2019). Meloscaphander grandis (Heterobranchia: Cephalaspidea), a deep-water species from the North Pacific: Redescription and taxonomic remarks. Zootaxa, 4646(2), 385–400. https://doi.org/10.11646/zootaxa.4646.2.12
- Dall WH. 1889 Reports on the results of dredging, under the supervision of Alexander Agassiz, in the Gulf of Mexico (1877-78) and in the Caribbean Sea (1879-80), by the U.S. Coast Survey steamer 'Blake'. Bull Mus Comp Zool 18 433–439
- Danovaro R, Company JB, Corinaldesi C, D’onghia G, Galil B, Gambi C, Gooday AJ, Lampadariou N, Luna GM, Morigi C, et al. 2010. Deep-sea biodiversity in the Mediterranean Sea: the known, the unknown, and the unknowable. PloS one. 5(8):e11832. doi:https://doi.org/10.1371/journal.pone.0011832.
- Darriba D, Taboada GL, Doallo R and Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat Methods, 9(8), 772–772. https://doi.org/10.1038/nmeth.2109
- Dautzenberg P and Fischer H. 1896. Dragages effectués par l’Hirondelle et par la Princesse Alice 1888–1895. 1. Mollusques Gastropodes. Mém Soc Zool Fr. 9:395–498.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi:https://doi.org/10.1093/nar/gkh340.
- Eilertsen MH and Malaquias MAE. 2013a. Systematic revision of the genus Scaphander (Gastropoda, Cephalaspidea) in the Atlantic Ocean, with a molecular phylogenetic hypothesis. Zool J Linn Soc. 167(3):389–429. doi:https://doi.org/10.1111/zoj.12013.
- Eilertsen MH and Malaquias MAE. 2013b. Unique digestive system, trophic specialization, and diversification in the deep-sea gastropod genus Scaphander. Biol J Linn Soc. 109(3):512–525. doi:https://doi.org/10.1111/bij.12069.
- Eilertsen MH and Malaquias MAE. 2015. Speciation in the dark: diversification and biogeography of the deep‐sea gastropod genus Scaphander in the Atlantic Ocean. J Biogeogr. 42(5):843–855. doi:https://doi.org/10.1111/jbi.12471.
- Etter RJ, Boyle EE, Glazier A, Jennings RM, Dutra E, Chase MR. 2011. Phylogeography of a pan-Atlantic abyssal protobranch bivalve: implications for evolution in the Deep Atlantic. Mol Ecol. 20(4):829–843. doi:https://doi.org/10.1111/j.1365-294X.2010.04978.x.
- Etter RJ and Grassle J. 1992. Patterns of species diversity in the deep sea as a function of sediment particle size diversity. Nature. 360(6404):576–578. doi:https://doi.org/10.1038/360576a0.
- Fischer P. 1883 Manuel de conchyliologie et de paléontologie conchyliologique VI (Paris)
- Grassle JF and Maciolek NJ. 1992. Deep-sea species richness: regional and local diversity estimates from quantitative bottom samples. Am Nat. 139(2):313–341. doi:https://doi.org/10.1086/285329.
- Holznagel W. 1998. Research note: a nondestructive method for cleaning gastropod radulae from frozen, alcohol-fixed, or dried material. Am Malacol Bull. 14(2):181–183.
- Huelsenbeck JP and Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17(8), 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
- Jobstvogt N, Hanley N, Hynes S, Kenter J and Witte U. 2014. Twenty thousand sterling under the sea: estimating the value of protecting deep-sea biodiversity. Ecol Econ. 97:10–19. doi:https://doi.org/10.1016/j.ecolecon.2013.10.019
- Kumar S, Stecher G, Li M, Knyaz C and Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution, 35(6), 1547–1549. https://doi.org/10.1093/molbev/msy096
- Locard A. 1897. Mollusques Testacés, tome premier. In: Milne-Edwards A, editor. Expédition scientifique du Travailleur et du Talisman pendant les années 1880, 1881, 1882, 1883. Paris: Masson et Cie; p. 1–516.
- Marcus EDR. 1974. On some Cephalaspidea (Gastropoda: Opisthobranchia) from the western and middle Atlantic warm waters. Bull Mar Sci. 24:300–371.
- Mighels JW and Adams CB. 1842 Description of twenty-four new species of New England shells J Nat Hist 4 37–54
- MolluscaBase. 2021, April 20. MolluscaBase. Scaphander Montfort, 1810. Accessed through: World Register of Marine Species. [accessed 2021 Apr 20]. http://www.marinespecies.org/aphia.php?p=taxdetails&id=137871.
- Montfort D. 1810 de Montfort PD. 1810. Conchyliologie systématique, et classification méthodique des coquilles (Paris)
- Mullineaux L, Speer K, Thurnherr A, Maltrud M and Vangriesheim A. 2002. Implications of cross-axis flow for larval dispersal along mid-ocean ridges. Cah Biol Mar. 43(3−4):281–284.
- Natural History Museum. 2014. Dataset: collection specimens. Resource: specimens. Natural History Museum Data Portal (data.nhm.ac.uk). doi:https://doi.org/10.5519/0002965.
- Pilsbry HA. 1893–1895. Polyplacophora. Acanthochitinidae, Cryptoplacidae and appendix. Tectibranchiata. Manual of Conchology, Structural and Systematic, with Illustrations of the Species 15, 181–436.
- Puillandre N, Lambert A, Brouillet S and Achaz G. 2012. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol. 21(8):1864–1877. doi:https://doi.org/10.1111/j.1365-294x.2011.05239.x.
- R Core Team. 2021. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. https://www.R-project.org/.
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA and Susko E. (2018). Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Systematic Biology, 67(5), 901–904. https://doi.org/10.1093/sysbio/syy032
- Sars GO. 1878 Bidrag til kundskaben om norges arktiske fauna: I. Mollusca regionis arcticae norvegiae, Christiania.
- Siegwald J, Pastorino G, Oskars T and Malaquias MAE. 2020. A new species of the deep-sea genus Scaphander (Gastropoda, Cephalaspidea) from the Mar del Plata submarine canyon off Argentina. Bull Mar Sci. 96(1):111–126. doi:https://doi.org/10.5343/bms.2019.0069.
- Thistle D. 2003. The deep-sea floor: an overview. Ecosystems of the deep oceans, 5.
- Tyler PA. 2003. Ecosystems of the Deep Oceans Tyler, PA ed. . In: Ecosystems of the world. Vol. 28. Amsterdam: Elsevier Science; p. 1–3.
- Van Dover C. 2000. The ecology of deep-sea hydrothermal vents. Princeton: Princeton University Press.
- Vogler RE. 2013. The radula of the extinct freshwater snail Aylacostoma stigmaticum (Caenogastropoda: Thiaridae) from Argentina and Paraguay. Malacologia. 56(1−2):329–332. doi:https://doi.org/10.4002/040.056.0221.
- Watson RB. 1883. Mollusca of the ‘challenger’ expedition. Part XX. J Linn Soc. 17:341–346.
- Watson GB. 1886. Report on the Scaphopoda and Gasteropoda collected by HMS Challenger during the years 1873-76. Reports of the Scientific Results of the Challenger Expedition, Zool., 42, 1–756.
- Wessel P and Smith WHF. 1996. A global, self-consistent, hierarchical, high-resolution shoreline database. J Geophys Res. 101(B4):8741–8743. doi:https://doi.org/10.1029/96JB00104.
- Zardus JD, Etter RJ, Chase MR, Rex MA and Boyle EE. 2006. Bathymetric and geographic population structure in the pan‐Atlantic deep‐sea bivalve Deminucula atacellana (Schenck, 1939). Mol Ecol. 15(3):639–651. doi:https://doi.org/10.1111/j.1365-294X.2005.02832.x.