ABSTRACT
During meiosis, both alleles of any given gene should have equal chances of being inherited by the progeny. There are a number of reasons why, however, this is not the case, with one of the most intriguing instances presenting itself as the phenomenon of meiotic drive. Genes that are capable of driving can manipulate the ratio of alleles among viable meiotic products so that they are inherited in more than half of them. In many cases, this effect is achieved by direct antagonistic interactions, where the driving allele inhibits or otherwise eliminates the alternative allele. In ascomycete fungi, meiotic products are packaged directly into ascospores; thus, the effect of meiotic drive has been given the nefarious moniker, “spore killing.” In recent years, many of the known spore killers have been elevated from mysterious phenotypes to well-described systems at genetic, genomic, and molecular levels. In this review, we describe the known diversity of spore killers and synthesize the varied pieces of data from each system into broader trends regarding genome architecture, mechanisms of resistance, the role of transposable elements, their effect on population dynamics, speciation and gene flow, and finally how they may be developed as synthetic drivers. We propose that spore killing is common, but that it is under-observed because of a lack of studies on natural populations. We encourage researchers to seek new spore killers to build on the knowledge that these remarkable genetic elements can teach us about meiotic drive, genomic conflict, and evolution more broadly.
INTRODUCTION
Functional meioses are all alike, but every dysfunctional meiosis is unique. Meiotic drive occurs when the sexual process is manipulated by given genes or haplotypes that, as selfish elements, can become overrepresented in the progeny. The transmission advantage of meiotic drivers may allow them to invade a population quickly, even if they impose a fitness cost to their host. An evolutionary arms race may ensue between host and driver, affecting genomic architecture and other aspects of evolution such as population divergence and demography (Burt and Trivers Citation2009). If a meiotic driver becomes fixed in a population, it becomes undetectable, as it can only drive in heterozygous states. Furthermore, the hallmark of meiotic drive, segregation distortion, is difficult to detect without intensive genetic investigations, unless it is linked to an obvious phenotype. Thus, the true prevalence of meiotic drivers in natural populations is unknown (Lindholm et al. Citation2016). Nevertheless, meiotic drivers have been identified in animals, plants, and fungi, with a great diversity of genetic architecture and modes of action (Lindholm et al. Citation2016). The focus of this review is on a group of meiotic drivers specific to ascomycete fungi for which new data are currently emerging at a high pace: the spore killers .
Table 1. Spore killer systems and their main characteristics
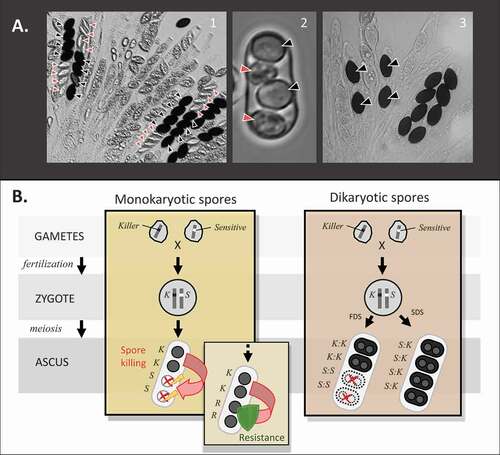
Fungi provide an ideal platform in which to study meiotic drive, as it can be directly observable due to the induced abortion of sibling spores. In ascomycete fungi, meiotic products are packaged together in individual sacs called asci. A given species will produce a set number of spores per ascus, usually four or eight. Meiotic drive can thus be observed by inspecting fruiting bodies for the presence of asci that possess only half the expected number of viable spores ().Like sperm or pollen killers found in animals and plants, a spore killer genetic element will kill meiotic products that do not carry the element and are sensitive to killing ().
Figure 1. General mechanism and observable phenotype of spore killing. A. Spore killing phenotype in three species of ascomycetes. A.1. N. sitophila (photo by Aaron Vogan). A.2. S. pombe (photo by Nicole Nuckolls). A.3. P. anserina (photo by S. Lorena Ament-Velasquez). Killer spores are indicated by black arrowheads and killed spores by red arrowheads, if visible. Notice that in S. pombe and N. sitophila, the aborted spores are small and colorless, whereas when spore killing is inflicted by Spok genes in P. anserina they disappear completely. B. Mechanism of spore killing in ascomycetes with monokaryotic or dikaryotic spores (see Glossary). Red arrows show the killing direction. Lightning bolts represent the killing action being effective on sensitive spores, whereas the green shield represents the killing action being ineffective on resistant spores. In zygotes heterozygous at the killing locus, sensitive spores (S) are eliminated while resistant spores (R) survive. In species with dikaryotic spores (e.g., P. anserina), killing only occurs among homoallelic spores resulting from first-division segregation (FDS), whereas heteroallelic spores resulting from second-division segregation (SDS) are protected.
Spore killers act through two main mechanisms: killer-target or poison-antidote (Bravo Núñez et al. Citation2018). In killer-target systems, the driver attacks a target element that is found in the genome of a sensitive individual, but not in the genome of the killer. In drivers that employ a poison-antidote mechanism, the poison that causes killing is produced together with an antidote, which protects the killer from suicidal action. In order to successfully drive, a target must never be inherited with a killer and the poison and antidote must always be inherited together. These mechanisms are effective in spore killing because after meiosis nuclei share a common cytoplasm, allowing products of one nucleus to diffuse to the other nuclei. After the formation of cell walls, the spores become isolated and experience the effects of the toxins. Killers with no targets, or which produce the antidote, are thereby spared the deadly fate of their sensitive sisters.
A number of repercussions may result from spore killing. As fungal spores are not gametes, but offspring, spore killing eliminates, in a simple case, half of the progeny of a mating event. However, in practice, the killing action will not necessarily result in halved fitness because of compensatory mechanisms. For example, if local sibling competition for resources is common among spores of the same ascus, spore killing could increase the survival probability of killer spores that eliminate their siblings, thereby leading to a partial recovery of the parent’s reproductive success (Lindholm et al. Citation2016). We refer to such putative positive fitness effects of killing as the killing advantage (see Glossary). Similarly, the parental mycelia may be able to recover some resources from aborted spores to produce additional progeny and replace a certain proportion of the killed offspring (Martinossi-Allibert et al. Citation2021; Vogan et al. Citation2021). Therefore, the fitness reduction to the host may be not as drastic as suggested by the intrinsic reduction in spore yield. This reflects a parallel with sperm and pollen killers, for which the reduction in gamete production does not necessarily translate into a reduction in offspring number (Hartl Citation1972).
Furthermore, the potential negative effects of spore killers can spur the evolution of resistance to killing (see Glossary), because strains that are able to suppress the action of the spore killer genes derive a fitness benefit. Alternatively, as the transmission of meiotic drivers relies on individuals reproducing sexually and outcrossing (e.g., Wright et al. Citation2008), affected populations may respond through modified mating strategies. Ascomycetes present a diversity of reproductive systems ranging from total asexuality to obligate outcrossing, including haploid and diploid selfing (Billiard et al. Citation2012). In many cases, outcrossing is a possibility but not a necessity (Nieuwenhuis and James Citation2016), which raises the question of the frequency of outcrossing events in ascomycetes and how that may relate to the transmission of spore killers.
Instances of spore killing have been found in numerous genera of ascomycetes: the model Sordariomycetes Podospora and Neurospora, the devastating plant pathogens Fusarium and Bipolaris, and the model fission yeast Schizosaccharomyces. We will briefly describe the characteristics of these known spore killers and their host species and then review important aspects of spore killers and their interactions with their hosts (summarized in ). First, we compare the genetic architecture of different spore killers and then examine resistance mechanisms. We follow with their associations with transposable elements (TEs), before summarizing theoretical knowledge of population dynamics of spore killers and their putative role in gene flow and speciation. We also discuss the possibility for future development of fungal synthetic drive based on naturally occurring spore killers. Lastly we put forth a call to arms for researchers across the field of mycology to examine their study species of choice for evidence of spore killing. Indeed, our knowledge of spore killers is rapidly growing but still limited. Fungal systems have much to tell us about how meiotic drive impacts evolution, and about genomic conflict from a broader perspective.
SPECIES
Podospora.—
Podospora anserina is a coprophilus species with a pseudohomothallic lifestyle (i.e., it packages two nuclei of opposite mating types into a single ascospore to ensure that offspring will be self-fertile upon germination; Raju and Perkins Citation1994). It has been used as a model system for genetics and molecular biology for much of the past century, and a wealth of wild collections exists, some of which date back to the 1930s. The very first spore killer elements were discovered in P. anserina (Bernet Citation1965; Padieu and Bernet Citation1967), although they were not identified as meiotic drive elements for a number of years (Turner and Perkins Citation1991; Dalstra et al. Citation2003). Subsequent genetic analyses of natural populations revealed upward of nine different spore killer types in populations of P. anserina (Van der Gaag et al. Citation2000; Hamann and Osiewacz Citation2004), some of which were assigned to Podospora spore killer designations Psk-1 through Psk-7 based on classical genetics (Van der Gaag et al. Citation2000). Later, a family of genes capable of spore killing was discovered. The first gene of this family, called Spok1 (for spore killing), was found in a strain of the sibling species P. comata. The second, Spok2, was identified in P. anserina itself (Grognet et al. Citation2014). At that point, the relationship between the Psk types and the Spok genes was unclear. More recently, high quality genomic sequencing of multiple Podospora spore killer strains and species revealed that three separate spore killer genes, Spok2, Spok3, and Spok4, are segregating at various frequencies within P. anserina (Vogan et al. Citation2019). Spok2 is present in nearly all strains investigated, except for a handful that appear to possess a deletion of Spok2, as indicated by a remnant and fragmented TE at the site (Vogan et al. Citation2019). This result suggests that the gene may have been fixed in the recent past. By contrast, Spok3 and Spok4 are typically found at low frequencies. These two genes entail a much more unusual case in that they occur within a large transposable element called Enterprise, forming a hyperselfish genetic element, the Spok block (Vogan et al. Citation2021). Additionally, it was found that Spok2, Spok3, and Spok4 have functionally diverged to provide no epistatic cross-resistance, so that strains with only Spok2 are susceptible to spore killing from Spok3 or Spok4 and strains with only Spok3 are susceptible to spore killing from Spok2 or Spok4, and so on. As a result, most strains of P. anserina are actually spore killers (Psk-S, containing only Spok2), some strains have a Spok block that can kill Psk-S, and a minority possess no spore killers at all (so-called “naive” strains). One strain has been reported that is resistant to Spok2, but its interactions with the other Spok genes is unknown (Grognet et al. Citation2014). Thus, through the presence and absence of the three genes, and multiple genomic locations of the Spok block with either Spok3 or Spok4 or both, a complicated hierarchy of spore killing is formed, which explains all previous observations of the different spore killing types (Vogan et al. Citation2019).
It is now apparent that P. anserina is part of a species complex that encompasses itself, P. comata, and five other species (Boucher et al. Citation2017). In the few P. comata strains that have been studied, Spok1 is always present at the same locus (Vogan et al. Citation2021). Intriguingly, Spok1 provides resistance against killing from all other Spok genes and is able to kill in the presence of Spok2 and Spok3, but not Spok4. Additionally, in another species of the complex, P. pauciseta, one strain has been discovered that possesses the Spok block with Spok3 and Spok4 (Vogan et al. Citation2019). Given that the individual species of the complex are extremely closely related (Vogan et al. Citation2019), yet seem to possess unique arrays of Spok genes, each lineage can be viewed as an independent natural experiment regarding the impact of meiotic drive on populations. Thus, understanding the dynamics of spore killing in this system will provide invaluable insights to the evolutionary role of meiotic drive on speciation (or lack thereof).
In addition to the Spok genes, there is another well-known spore killer in P. anserina, het-s (Dalstra et al. Citation2003). There are two alleles of the het-s gene: het-s and het-S. The HET-s protein product exists as a prion, which when it encounters a cell of het-S genotype induces a cell death response. This prion-induced cell death occurs in both vegetative and sexual tissues. In the vegetative state, cell death will prevent anastomosis between two strains with opposite het-s alleles, a phenomenon known in fungi as heterokaryon incompatibility, and well characterized in P. anserina (Coustou et al. Citation1997; Pinan-Lucarré et al. Citation2007). In the sexual cycle, the cell death reaction occurs when the maternal strain is HET-s prion infected and the paternal strain is het-S, resulting in segregation distortion toward the het-s genotype. The reaction is temperature sensitive, however, and has been observed at 18 C, but not at 25–28 C (Bernet Citation1965; Dalstra et al. Citation2003). The molecular mechanism of how the two protein products interact to result in meiotic drive is perhaps the best understood of any known meiotic drivers (Riek and Saupe Citation2016), but given the specific circumstances that are required for the drive to occur, and its dual role as a heterokaryon incompatibility gene, it is unclear how important the driving behavior is in nature. As with other heterokaryon incompatibility genes, het-s is likely subject to balancing selection forces, which should lead to approximately balanced allele frequencies (Debets et al. Citation2012; Milgroom et al. Citation2018). Still, the het-s allele was found to be overrepresented in a monitored population, consistent with an effect of drive in its dynamics (Debets et al. Citation2012).
Neurospora.—
Neurospora is relatively closely related to Podospora (divergence time, ~100 million years ago [mya]; Lutzoni et al. Citation2018), and it has been used as a model for as long. Neurospora crassa is heterothallic and has remained one of the most important fungal model systems. However, it is within the closely related species, N. sitophila and N. intermedia, where the spore killers have been found. Together, these two species harbor three known spore killers: Sk-1, Sk-2, and Sk-3 (Turner and Perkins Citation1979). The latter two are the best studied, as they occur in the species N. intermedia and have been introgressed into N. crassa for genetic characterization under laboratory conditions (Campbell and Turner Citation1987). Sk-1 is found in N. sitophila and was only described in depth recently (Svedberg et al. Citation2021).
Throughout the years, thousands of Neurospora isolates have been collected from around the world and phenotyped for the presence of the spore killers. Sk-1 is prevalent in ~15% of N. sitophila isolates but shows a patchy distribution, appearing fixed, absent, or polymorphic at given locales. Conversely, Sk-2 has only been found in four isolates and Sk-3 in just one. However, resistance phenotypes are commonly found for both Sk-2 and Sk-3, with some strains exhibiting resistance to both (Turner Citation2001). For Sk-1, only a single isolate was ever found to show resistance, but population genetic analyses suggest that the resistant strain may belong to a distinct lineage or a yet undescribed species; therefore, the prevalence of resistance to Sk-1 in nature is still obscure (Svedberg et al. Citation2021).
The Sk-1 locus was only recently described at the genetic level. Spore killing by Sk-1 strains is caused by a single gene, Spk-1, that resides on chromosome 6. So far, little is known about Spk-1, but phylogenetic analyses suggest that it may have been introgressed to N. sitophila from another Neurospora species, N. hispaniola. Additionally, numerous potential homologs appear to be scattered across the genomes of many strains and species of Neurospora, suggesting that it could be a member of a large gene family, although none of the other homologs have known function (Svedberg et al. Citation2021). Although no specific resistance mechanisms against Spk-1 have been identified, the genome defense machinery known as meiotic silencing of unpaired DNA (described in Mechanisms of Resistance), is able to suppress activity of Spk-1 under certain circumstances. The ability of strains to suppress Spk-1 appears to be population specific, and future studies may reveal important evolutionary dynamics between Sk-1 and host genome defense.
The Sk-2 and Sk-3 killer loci are distinguished from each other based on mutual killing. That is, when an Sk-2 strain is crossed to an Sk-3 strain, no viable spores are observed. Both spore killers encompass large haplotypes of ~400 genes and have separate killing and resistance loci linked by a nonrecombining region (Campbell and Turner Citation1987; Svedberg et al. Citation2018). This is akin to well-described sperm killers from mice and flies (Lindholm et al. Citation2016), but unlike all other known spore killers. The two haplotypes span a similar region, straddling the centromere of chromosome 3, and share the same resistance locus (called rsk) (Hammond et al. Citation2012). The killer locus of Sk-2 has been identified (rfk), but there is no homologous gene in the Sk-3 region (Harvey et al. Citation2014; Rhoades et al. Citation2019). Additionally, a comparison of the spore killer regions shows significant divergence, including independent inversions (Svedberg et al. Citation2018). Additional observations suggest that separate genetic incompatibilities have been accumulating within Sk-2 and Sk-3 that increase the reproductive boundary between N. intermedia and its sister species, N. metzenbergii (Vogan et al. Citation2020), consistent with a protracted period of independent evolution between the killers. Interestingly, for both Spk-1 and rfk (of Sk-2), RNA A-to-I editing occurs, which may either be a general feature of meiotic drive genes or a particularity of how Neurospora controls its sexual cycle.
Schizosaccharomyces.—
The model fission yeast, Schizosaccharomyces pombe, is the only species outside of the Pezizomycotina in which spore killers have been found. Although S. pombe is extremely well studied in the laboratory, its natural history is poorly understood. Additionally, strains of S. pombe are capable of mating type switching, a process that allows clones to mate among themselves (i.e., to go through haploid selfing). Thus, it is unclear how often mating among unrelated individuals occurs in the wild. Although the spore killing in Schizosaccharomyces pombe was identified most recently (Zanders et al. Citation2014), an impressive amount of insight has been gleaned in a short span of time. In this system, spore killing was first identified between the standard laboratory strain and a strain isolated from kombucha that is sometimes treated as a separate species but now known to be a hybrid between two divergent ancestral populations (Tusso et al. Citation2019). The genes responsible for drive in S. pombe belong to the wtf gene family (referring to “with Tf”; see below, Spore Killers and Their Romance with TEs), which can be present in dozens of copies per strain (Hu et al. Citation2017; Nuckolls et al. Citation2017). These are single-gene drivers that encode both a poison and an antidote through alternative splicing. The full open reading frame (ORF) expresses the poison, whereas a splice variant that uses a downstream start codon that excludes the first exon codes for the antidote. Multiple wtf genes have been identified from global collections of S. pombe, including those that lack the upstream exon and thus only code for the antidote but not the poison (i.e., they act as resistance alleles; see below, Mechanisms of Resistance) (Núñez et al. Citation2018). Due to individual differences in copy numbers between strains, the wtf genes contribute significantly to low spore viability in crosses between even closely related strains, through mutual killing in S. pombe (Eickbush et al. Citation2019; Bravo Núñez et al. Citation2020a, Citation2020b). The exact details of the molecular mechanism through which the wtf genes act is not fully understood, but initial results suggest that it may involve protein aggregation, as in the case with het-s in Podospora (Coustou et al. Citation1997; Nuckolls et al. Citation2020).
Fusarium.—
Fusarium is a cosmopolitan group of plant pathogens that spans some 70+ million years of divergence (Lutzoni et al. Citation2018). Within the genus, many species have no known sexual cycle, although sex is ancestral to the genus (Ma et al. Citation2013). Fusarium can further be divided into a number of complexes that differ in host plant specificity (Summerell Citation2019). At least two separate spore killers have been identified from Fusarium. One spore killing locus has been identified from Fusarium verticillioides (= Fusarium moniliforme). Up to 80% of wild isolates carry a spore killing locus, which drives with an efficiency of ~95%, although some isolates appear to be partially resistant (34%–61% killing efficiency) (Kathariou and Spieth Citation1982). Later efforts were successful in locating the spore killer locus to a 102 kb region on chromosome 5 of F. verticillioides, but the exact genetic underpinnings are currently unknown. Analysis of the region did identify three hypervariable regions, which may harbor the spore killer genes (Pyle et al. Citation2016). This finding suggests that the Fusarium spore killer could be a multilocus system like in Neurospora Sk-2/3, although it would have to be significantly smaller.
Another locus has been identified in F. fujikuori (= Gibberella fujikuroi), which is part of a large species complex, formerly referred to by multiple “mating groups” (O’Donnell et al. Citation2000). A follow-up investigation of wild strains of F. subglutinans (= Gibberella fujikuroi var. subglutinans) confirmed that this second spore killer was also present in this clade (Sidhu Citation1984) and thus may be widespread in the complex.
Bipolaris.—
The most understudied spore killer was identified in Bipolaris maydis (= Cochliobolus heterostrophus), a dothideomycete pathogen of corn. It was found that the locus responsible for production of the T-toxin (an important compound for plant virulence) was tightly linked to a meiotic drive locus, such that some strains that do not produce the T-toxin (also called race O) contain a killer locus; all T-toxin–producing strains (race T) appear to be sensitive to killing (Taga et al. Citation1985). The story is complicated by the fact that the T-toxin locus is involved in a chromosomal rearrangement that also results in abortive ascospores, but this alone does not account for the observed drive (Bronson Citation1988, Citation1990). The waters were muddied further when it was discovered that the T-toxin locus was actually composed of two separate loci on different chromosomes, such that the relative location of the spore killer locus became ambiguous (Turgeon et al. Citation1995; Turgeon and Baker Citation2007). To our knowledge, further efforts to characterize the spore killer locus have so far not been undertaken, so the details of the drive remain virtually unknown.
GENOMIC ARCHITECTURE OF SPORE KILLERS
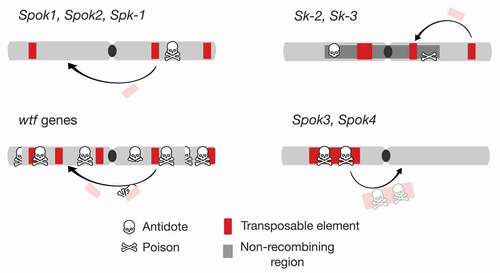
Like all killer meiotic drivers, spore killers must perform two key functions in order to cause drive: they need to (i) destroy the meiotic products that do not inherit the drive locus and (ii) at the same time protect themselves. Close genetic linkage between the two factors ensures that the two elements do not become uncoupled and create a situation where the spore killer destroys itself. Although linking two separate genes with distinct functions could be accomplished by having them in close physical proximity, the general expectation is rather that genomic rearrangements will evolve that encompass both genes and prevent recombination from separating them. Sex chromosomes provide an attractive target for the evolution of these systems, as heteromorphic chromosomes will already have large nonrecombining regions, which could serve as hot spots for the origin of meiotic drive genes (Frank Citation1991a; Hurst and Pomiankowski Citation1991; Jaenike Citation2001). In the case of the spore killers, however, the norm seems to be single-gene systems ()
Figure 2. Genome architecture of spore killers. Within a genome, represented by a schematic chromosome (drawn as a gray bar with black centromere), spore killers (symbol: skull and bones) are found as either single genes with both the antidote and poison functions (most known instances, e.g., Spok1, Spok2, and Spk-1) or as multigene systems within nonrecombining regions (Sk-2 and Sk-3). In some cases, the spore killer genes are in low copy number and can be associated with TEs (e.g., Spok1, Spok2) or not (e.g., Spk-1). In the case of the wtf gene family, TE-associated mobilization and other processes result in the proliferation of multiple copies, including those with only a functional antidote or that are fully pseudogenized. As a special case, the Spok3 and Spok4 genes are only found within a large TE (Enterprise), which relocates them during transposition.
Still, there could be factors that make multigene complexes less likely to evolve in fungi. One potential difference lies in the consequences of structural rearrangements such as inversions. In fungi, large inversions can often be inferred through a significant production of inviable spores in crosses heterozygous for the inversion (Perkins Citation1974), which can be expected to have significant negative fitness effects. In many other organisms, such as Drosophila, several mechanisms are present that will reduce these fitness effects and segregating inversions are therefore more tolerated and relatively common (Reis et al. Citation2018). In such cases, it is easy to see that inversions can suppress recombination locally on a chromosome, but if inversions are instead accompanied by a reduced reproductive output, they may be less likely to provide a fitness increase. Sk-2 and Sk-3 carry inversions spanning a 30–40 cM region (~1.8 Mb) (Campbell and Turner Citation1987; Svedberg et al. Citation2018), but despite this, a corresponding elevated production of inviable spores beyond what is caused by the spore killing itself has not been observed (Turner and Perkins Citation1979). This may be explained by the presence of other mechanisms that can reduce recombination, as has been observed in N. tetrasperma (Sun et al. Citation2017), but little is still known about these mechanisms and to what extent they can facilitate the proliferation of multigene complexes. Additionally, little is known about the effects of genetic background on recombination rates, especially since much of the genetic analysis of Sk-2 and Sk-3 was performed in an N. crassa background (Turner and Perkins Citation1979). Further work is thus necessary to determine how they affect the observed recombination patterns.
Multigene complexes not only can facilitate meiotic drive by linking toxin and antidote genes, but sometimes they will also contain other genes that have evolved to increase the efficiency of the drive (Crow Citation1991b; Hurst and Pomiankowski Citation1991). Such genes are called “enhancers,” and although some have been identified in other meiotic drive systems (Charron et al. Citation2019; Courret et al. Citation2019), none have been observed in spore killers, with the possible exception that the Sk-2 and Sk-3 complexes may carry a factor that blocks the genome defense system MSUD (see Genome Defense, below). In the cases of single-gene spore killers, these might provide less of a substrate to evolve tight linkage to, and one might also speculate that single-gene drivers could be less deleterious on average and thus fix more readily, leaving little time for enhancers to evolve. Furthermore, most spore killers appear to be highly efficient, and there may simply be little need for additional factors that can increase killing efficiency.
Although at first glance single-gene drivers may appear to be simple in their structure and organization, the reality is quite the opposite. A number of gene duplications are involved with both the Spok and wtf genes, and putatively for Spk-1 (Svedberg et al. Citation2021), leading to gene family expansion. In Podospora anserina, this has created at least three separate copies: Spok2, Spok3, and Spok4 (Vogan et al. Citation2019). Additionally, a pseudogenized Spok gene has also been identified, which could be descended from either Spok3 or Spok4, as it occurs in a degraded Spok block (Vogan et al. Citation2021). Alternatively, it may be another independent homolog. In S. pombe, the wtf genes have expanded to an even greater extent, such that some strains possess as many as 38 copies (Bravo Núñez et al. Citation2020a) and the total number of homologs in the species could be larger (Eickbush et al. Citation2019). Immediately after a duplication event, the new spore killing gene will have no effect on drive dynamics if it is linked to the original copy, but it can drive if it locates to an unlinked locus due to recombination and independent assortment during meiosis. However, the spore killer genes can subsequently diversify in a number of ways, each with unique effects on the dynamics of spore killing. First, they may evolve into unique drivers, with no epistatic interactions. Second, they may lose the ability to kill but retain their antidote function, effectively becoming a resistance allele (see below). Third, a hierarchy may form whereby one homolog can drive in the presence of another, but not vice versa (e.g., Spok1 vs. Spok2). Lastly, the new homolog may degrade and lose all functionality but could still activate genome defense machinery. Thus, although single-locus drives may originate as simple systems, they may become an interlinked network of complex interactions involving competing drivers, and suppression. Therefore, one may question whether the known array of multigene drivers all originated as such, or conversely, whether some may have begun as single-gene drivers that subsequently expanded to additional loci, connected by genomic rearrangements.
MECHANISMS OF RESISTANCE
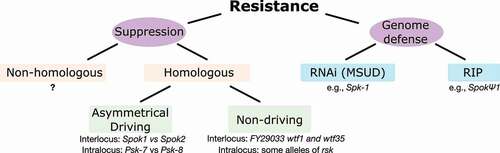
The potential fitness costs of spore killers, caused by the destruction of spores, genomic inversions, or hitchhiking deleterious mutations, create a selective pressure that favors mechanisms counteracting or disabling the drive. We propose to distinguish the action of “suppressors” (see Glossary), factors that specifically target spore killers, from generalist genome defense mechanisms that can also affect spore killers under specific circumstances but are not restricted to that action (). In general, selfish elements are dependent on their genomic context; thus, we do not consider cases where spore killers fail to function in novel populations (as may be the case for Sk-1; see Neurospora section) as resistance. Because of their specificity, suppressors are generally expected to invade a population in response to a costly spore killer, whereas genome defense mechanisms likely exist in the population before the invasion of the driver. In many instances, the population dynamics of the host and driver likely determine the fate of the driver, as selection will act to purge the driver from the population if it is too costly. However, in the context of genome defense, there may exist a more nuanced game of “cat and mouse,” with successful drivers being the ones that avoid detection by the host in the first place.
Figure 3. Mechanisms of resistance. The phenotype of resistance (partially or completely avoiding spore killing) can occur either due to the action of specific genetic elements (suppressors) or to general genome defense mechanisms. All known suppressors are homologous to their corresponding spore killer elements, although in principle nonhomologous suppressors could also evolve. In turn, homologous suppressors could be restricted to the antidote function and thus unable to drive, or be drivers themselves, creating asymmetrical killing relationships. Some known examples are given. Within fungi, two genome defense mechanisms are known to deactivate meiotic drivers: MSUD (e.g., working against Spk-1) and RIP (e.g., as hinted by the pseudogenized SpokΨ1 gene).
Suppressors.—
In the context of spore killing, suppressors are genes that lead to a phenotype of resistance, where the loss of viable spores is reduced or absent. In all known examples, suppressors are homologous to the antidote function of the killer that they confer resistance to, although in theory this is not the only possible mechanism. Below, we distinguish different classes of suppressor genes, with different evolutionary origins, leading to different expressions of the resistance phenotype (see ).
A first scenario, for which several examples are known, is the case of a sequence homologous to the killer that lacks the ability to kill spores but carries an active antidote function, thus leading to a resistant phenotype. Such homologous suppressors can arise from several mechanisms depending on the genomic architecture of the driver. For spore killers that operate with separate poison and antidote genes (or killer and target genes), the association between the two loci may be less strict than in the case of single-gene killers. Rare recombination events may thus give rise to nondriving homologous suppressors by separating the two components. For such two-gene systems, an alternative scenario must also be considered, whereby resistance is ancestral to the population: when first arising, a killer gene could co-opt an antidote function from a particular allele that is polymorphic in the present population (Sweigart et al. Citation2019). If that is the case, an allele with the antidote function already exists in the population and represents a homologous nondriving suppressor (). Although this scenario involving a preexisting antidote may appear counterintuitive, it is to our knowledge the most likely explanation for the well-known rsk suppressors found in N. intermedia, where resistance is common in Southeast Asia, the same locale in which the Sk-2 and Sk-3 spore killers were found (Turner Citation2001). The rsk gene, which corresponds to the antidote function of Sk-2 and Sk-3, also exists at the same locus in some non-killer strains and acts as a suppressor, with different alleles conferring resistance to Sk-2, Sk-3, or both. The different rsk alleles show high degrees of sequence divergence (up to 20%) within N. intermedia, suggesting that they were segregating in the species before the appearance of Sk-2 and Sk-3, and it is therefore possible that the spore killer precursor genes captured already existing alleles, which allowed them to suppress their own killing (Svedberg et al. Citation2018). In this case, resistant rsk alleles may have spread locally as a consequence of the presence of spore killers in the region, but it is also possible that an initial higher frequency of resistant alleles due to some other reason made it more likely for spore killers to evolve in that geographic area.
In the case of single-gene drivers such as the Spok genes and wtf genes, homologous suppressors can arise by loss of the killing function. In S. pombe, in which the mechanism of drive relies on an alternative start codon, loss of the upstream start site will prevent the transcription of the toxin but still allow the antidote to be produced (Hu et al. Citation2017; Nuckolls et al. Citation2017). In Podospora, although a single protein enacts both functions, the mutation of a single residue can halt toxicity, resulting in an allele only conferring resistance (Vogan et al. Citation2019). Thus, it may be that for poison-antidote systems, the poison function can generally be separated from the antidote function to produce a nondriving suppressor (i.e., a resistance allele). Whether this suppressor invades and leads to extinction of the driver will depend on a number of factors, in particular the fitness cost imposed by the killer. Consequently, even though suppressor alleles may be likely to be produced, they may never reach appreciable frequencies in the population.
A second scenario of resistance, probably more restricted, involves homologous sequences at a different locus that retain the ability to drive but are able to kill one another in a nonreciprocal fashion (). This type of asymmetrical driving leads to instances where some killers confer resistance to others. This scenario is observed to be the result of gene family expansion and is observed for both the Spok and wtf genes. Among the Spok genes, Spok2, Spok3, and Spok4 have rapidly diverged functionally, so as to be completely independent drivers. However, Spok1 is able to suppress the killing of all Spoks and is only suppressed by Spok4. The wtf genes have numerous variants that have lost the killing action and thus serve as resistance loci for various drivers, but there are also instances (e.g., FY29033 wtf1 and FY29033 wtf35) where apparent gene conversion between wtf genes results in copies that, while still functioning as independent drivers, acquire the ability to suppress other wtf genes (Eickbush et al. Citation2019; Bravo Núñez et al. Citation2020a). Although we currently only have two examples of spore killing gene family expansion, they both suggest that the prevailing forces promote diversification into independent drivers, rather than maintaining epistatic interactions for suppression. With Podospora, only Spok1 has suppressive effects, and it is found in a separate species, P. comata, indicating that it has not directly evolved with the influence of the other Spok genes. In S. pombe, the large number of copies of wtf genes makes it difficult to understand their specific interactions, but given the large degree of mutual killing between strains, many copies must have unique killing functions (Zanders et al. Citation2014). However, as S. pombe appears to outcross rarely, it may be the case that drive occurs rarely in nature and thus that many of the factors affecting wtf gene diversity are incidental (but see Hernández et al. Citation2021).
We have seen that homologous suppressors can arise by loss of the killing function in both one- and two-gene spore killers. This type of suppressor can then occur at the same locus as the killer itself (intralocus homologous suppressor; see ), but also at a different locus in the case of killers undergoing gene family expansion, such as the wtf and Spok genes (interlocus homologous suppressor; see ). If the spore killer and its suppressor are found at different loci (unlinked), this can have profound effects on the ability of a spore killer to spread, and the selective pressures to evolve new specificities that can evade existing suppressors, but little is known about these dynamics. In S. pombe, interlocus suppression has been observed (Bravo Núñez et al. Citation2020a), but more work remains to be done to fully understand this complex meiotic drive system.
Finally, it is in theory possible that resistance appears because of a nonhomologous gene, likely unlinked, that, for example, produces an inhibitor to the poison without homology to the antidote function of the killer itself. However, examples of nonhomologous suppressors are not yet known in spore killers. It may therefore be unlikely to evolve in practice.
Genome defense.—
In ascomycete fungi, two primary forms of genome defense may prevent the invasion of spore killers: repeat induced point mutation (RIP) affecting multicopy killers and RNAi-mediated silencing known as meiotic silencing of unpaired DNA (MSUD) (). Both phenomena are active during the sexual cycle and could prevent spore killers from functioning.
RIP is one of the most prevalent genome defense mechanisms across ascomycetes and is easily detectable on the genomic scale in many species (Clutterbuck Citation2011). It operates by targeting any duplicate sequence within the genome and induces C-to-T mutations, often within a specific di- or trinucleotide context (Cambareri et al. Citation1989). It appears to be widespread across Pezizomycotina, but basidiomycetes may possess analogous machinery to mutate repetitive regions (Horns et al. Citation2012). RIP will target genes just as effectively as selfish genetic elements such as TEs or meiotic drivers, and so it may come at a cost of limiting innovation within genomes (Catalanotto et al. Citation2006). RIP is very well studied within Neurospora, where it was discovered, and functions in P. anserina (Graïa et al. Citation2001) and Fusarium solani (= Nectria haematococca) (Coleman et al. Citation2009), among others, but it is absent from S. pombe. As RIP is a stochastic process that targets both copies of a duplicate sequence, it can spur the divergence rate between paralogs. Although the majority of mutations are likely to be deleterious and result in nonfunctional gene products, some will likely still be functional and may even result in a shift in poison-antitoxin interactions to derive novel drivers. This action could explain, for instance, the rapid divergence of Spok3 and Spok4 in Podospora, which is likely the result of a tandem duplication, but they now display no cross-resistance or other epistatic interactions (Vogan et al. Citation2019). RIP has also presumably been successful at eliminating at least one Spok block (and associated Spok gene) from the population, as evident from a degraded copy that is polymorphic within P. anserina (Vogan et al. Citation2021). RIP therefore has a complex interaction with spore killers wherein it will be entirely ineffective against single-copy spore killers but then either enact the destruction or promote the diversification of multicopy drivers.
The RNA silencing machinery of fungi is homologous to that from plants and animals (Cerutti and Casas-Mollano Citation2006), and so direct parallels between fungi and the well-investigated drivers in Drosophila can be made here. Although S. pombe has RNAi, it appears to be primarily used for heterochromatin formation and has little impact on the transcription of TEs or the wtf genes (Hansen et al. Citation2005). On the other hand, RNAi has profound effects on TEs in Neurospora, through the MSUD pathway. Unlike RNAi in Drosophila, which targets duplicated sequences, the MSUD machinery silences RNA expression of any genomic sequence that has no homolog on the sister chromatid during meiosis. This includes transgenes, which led to the discovery of MSUD (Shiu et al. Citation2001). As a result, any novel insertions or indels in heteroallelic crosses will be silenced. This should mean that a spore killer that is present in one strain but not the other will not be transcribed and thus should be ineffective in Neurospora. However, all three spore killers in Neurospora evade MSUD to various extents. For Sk-2 and Sk-3, it has been hypothesized that the large amount of unpaired sequence in the spore killer haplotype overwhelms the MSUD machinery and allows the drive to function (Raju et al. Citation2007). In N. sitophila, the situation is more complex. Most of the time, the Sk-1 driver seems to avoid being silenced through an unknown process. However, some populations of N. sitophila are able to mount a successful MSUD response to inhibit the action of the drive (Svedberg et al. Citation2021). Efficiency of MSUD increases with the relatedness of the cross, but exactly why some strains can silence the drive while others cannot will be the subject of future research.
Studies of genome defense are still relatively restricted to a handful of model systems. RIP appears to be widespread in ascomycetes, and although the machinery for RNAi production is well conserved across fungi (Choi et al. Citation2014), MSUD has only been described in a limited number of species (Nakayashiki et al. Citation2006; Son et al. Citation2011). These observations on different types of suppressors paint a complex picture whereby the propensity for a spore killer to successfully invade depends not only on the intrinsic properties of the driver, but also on the genomic background within which it evolves, highlighting the need to gather data on drivers in natural populations. Studying the interaction between meiotic drive and genome defense has revealed a great deal of information about how both systems function. An important question for the future will be to discover how universal these dynamics are across systems and species, or whether given meiotic drivers will be shown to have unique interactions with their host’s defense machinery.
SPORE KILLERS AND THEIR ROMANCE WITH TEs
In both Podospora and S. pombe, the spore killer genes have conspicuous associations with transposable elements (TEs; ). As mentioned above, the name wtf is short for “with Tf,” a retrotransposon that is commonly found flanking the wtf genes (Bowen et al. Citation2003). All four Spok genes are either flanked by TEs (Grognet et al. Citation2014) or are found within the giant transposon Enterprise (Vogan et al. Citation2021). The wtfs were first identified due to their large copy number and TE association, long before their role as spore killers was known. Long terminal repeat (LTR) elements are particularly prone to ectopic recombination, and the wtf genes are in double-stranded DNA breakage hot spots (Eickbush et al. Citation2019), suggesting that TEs may help drive diversification of the gene family. Spok1 and Spok2 are found within TE-rich regions in P. comata and P. anserina, respectively. These regions are both located on the same arm of chromosome 5, but at different locations (Grognet et al. Citation2014). Although both Spoks are seemingly in high frequency, and all species within the P. anserina complex are extremely closely related, the corresponding locus of each Spok shows no indications of past Spok genes in the other species. Additionally, a third species, P. pauciseta, also shows no traces of Spok genes in these regions (Vogan et al. Citation2019). As the genomes of these species are highly syntenic, once again ectopic recombination induced by TEs appears likely for moving the Spok genes among loci. Spok3 and Spok4 are unique in that they are actively moving through the genome because they have been captured by Enterprise. Furthermore, the two homologs may have arisen as part of a duplication within Enterprise (Vogan et al. Citation2021), hinting that the size and/or composition of this TE is playing a role in the diversification of the Spok genes. Both the wtf genes and the Spok genes are also known to undergo gene conversion between copies (Eickbush et al. Citation2019; Vogan et al. Citation2019), entangling their evolutionary relationships and making it difficult to precisely reconstruct past events.
In N. sitophila, Spk-1 is not found in direct association with TEs. However, the putative ORF responsible for drive appears to have been inserted into another gene, truncating it. Additionally, numerous homologs of Spk-1 can be found distributed across the genomes of the various species of Neurospora, suggesting that they too are somehow being translocated. Although N. crassa has little to no direct evidence of active TEs, variation in TE content between closely related strains and species of Neurospora suggests that transposons are not static across the genus (Nguyen et al. Citation2020). Given that only Spk-1 has been identified as a spore killer, the other copies may no longer be capable of driving, and what we observe is a landscape of dead copies. Perhaps in controlling its TE content, Neurospora also ended the proliferation of this gene family, ultimately resulting in the near extinction of this driver. The Sk-2/Sk-3 locus is also enriched for TEs, but given its size, it is unknown whether these TEs played a specific role in its evolution as a spore killer, or whether they simply accumulated there due to lack of purifying selection. The latter option is supported by the fact that the region shows a signature of accumulation of slightly deleterious mutations (Svedberg et al. Citation2018).
Although the fate of a successful spore killer is fixation, there could be ample time during this process, or even after, in which a spore killer could find itself in another genomic location. If the translocation is due to an associated TE, then the spore killer may become permanently associated with it as the cycle of drive–fixation–translocation–drive repeats itself. However, if the movement of the spore killer occurs at a higher rate than fixation, it may never fix but rather exist in polymorphic states at various genomic loci. This could be the case for both Spok3 and Spok4 within Enterprise, as well as with many wtfs in unstable genomic regions. This may ultimately have disastrous consequences on reproduction, as when the spore killers ultimately diversify into independent drivers, heterozygous crosses will lead to mutual killing and may result in low outcrossing viability, as observed in S. pombe. In the end, this could either lead to selection removing the driver altogether or result in a drastic shift in mating system or population structure. Studying the evolution of selfing vs. outcrossing rates in Podospora and S. pombe could potentially reveal underappreciated roles of meiotic drive in shaping population dynamics. Conversely, different populations could fix spore killers at separate loci while diverging. If these populations eventually undergo speciation, then the drivers could contribute to reproductive isolation between the species.
SPECIATION AND GENE FLOW
New meiotic drivers are often discovered in hybrid crosses between different species or populations (Burt and Trivers Citation2009; Bravo Núñez et al. Citation2018). In that context, meiotic drive can be seen as a type of intrinsic postzygotic isolation barrier with a potential role in the speciation process (Coyne and Orr Citation2004; Frank Citation1991b; Hurst and Pomiankowski Citation1991; Lindholm et al. Citation2016). As meiotic drivers can go to fixation faster than neutral variants, in principle any associated reproductive incompatibilities can also build faster between diverging populations. In animals and plants, the effects of meiotic drive in reproductive isolation are most often discussed in relation to sex chromosomes (Haldane’s rule) and associated modifiers (suppressors and enhancers), or to female centromeric drive (Frank Citation1991a; Hurst and Pomiankowski Citation1991; Coyne and Orr Citation2004), although there are certainly other types of drivers with fertility effects (Zanders and Unckless Citation2019). As fungi have no sex chromosomes, and production of male and female reproductive structures (e.g., trichogyne and spermatia, equivalent to gametes) is decoupled from meiosis in the life cycle, we can study the role of autosomal drivers in the speciation process more directly. Here, we distinguish three particular models of spore killing effects on reproductive isolation, all starting from two allopatric populations.
In the first model, a spore killer invades one of the populations and drives to fixation ()
Figure 4. Models of spore killing effects on reproductive isolation. Starting from two idealized populations that diverge in allopatry, spore killers can have different effects on the buildup of reproductive barriers. A. A spore killer invades a population and a genomic conflict ensues. Along with suppressors and enhancers, other mutations can evolve and increase in frequency, acting as Bateson-Dobzhansky-Muller (BDM) incompatibilities if the invaded population comes into contact with a naive population. As a result, the hybrid crosses have additional incompatibilities on top of spore killing. B. Both populations accumulate different spore killers. Upon hybridization, the resulting spores are killed at various degrees depending on linkage between the spore killers, which lowers hybrid fitness and creates a reproductive barrier. C. Finally, a spore killer accumulates in one population, and like in A, it causes spore killing in the hybrid crosses. However, in this scenario, the spore killer eventually invades the naive population, lowering average divergence and hence reducing potential incompatibilities between the populations.
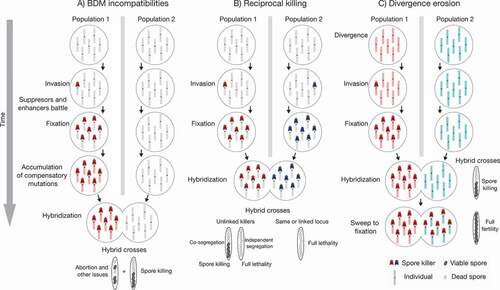
As is probably the case with Sk-2 and Sk-3, the BDM incompatibilities associated with the spore killer haplotype might not be the original cause of speciation. Instead, they might act as additional reproductive barriers between already diverged species. However, the spore killers can theoretically lead to a peculiar situation where their selfish action results in an extreme case of postzygotic isolation, even in the absence of other incompatibilities (). If two noninteracting spore killers fix independently in allopatric populations, hybrids would be the victim of the action of two poisons upon secondary contact (Bravo Núñez et al. Citation2018). If the spore killers are unlinked, they could either segregate together randomly, producing spore killing, or segregate independently, in which case all spores (i.e., all hybrids) would die. At the extreme, full hybrid lethality would occur if the two killers are located at the same locus. We call this second model the “reciprocal killing” reproductive barrier. Such a lethal cross has been artificially engineered by putting either Spok3 or Spok4 at the same locus in P. anserina (Vogan et al. Citation2019) and is observed when crossing Sk-2 and Sk-3 Neurospora strains (Turner and Perkins Citation1991). At the same time, as more spore killers accumulate in each population, the effects of reciprocal killing get multiplied, resulting also in extremely low viability even if the killers are unlinked. For example, there are so many wtf genes in different S. pombe strains that crosses are often largely sterile (Zanders et al. Citation2014; Hu et al. Citation2017; Nuckolls et al. Citation2017). At this point, however, the extent of reciprocal killing in natural populations of S. pombe is unknown, and on the contrary, genetic background may predominantly determine spore viability (Tusso et al. Citation2019).
The third model highlights the fact that the selfish nature of the spore killers might go against, instead of in favor of, speciation (). As a population fixed for one spore killer comes into contact with a naïve population, the segregation distortion advantage of the killer might in fact promote the invasion of the spore killer into the new population. Thus, the spore killer might sweep quickly in the new background, bringing with it linked variants from the other population. In this case, the divergence between the two populations decreases, potentially removing other genetic incompatibilities (Crespi and Nosil Citation2013). This situation has been famously observed in Drosophila (Meiklejohn et al. Citation2018). Similarly, it is known that, in spite of strong postzygotic isolation in the laboratory, the region containing Sk-1 in N. sitophila was introgressed from another species, N. hispaniola (Svedberg et al. Citation2021). However, as little is known about the population dynamics of most spore killers and their host species, it remains unclear just how much spore killers transit through different fungal populations. Naturally, what could start as a reciprocal-killing situation could later turn into divergence erosion if the few surviving offspring are enough for the spore killers to persist and invade the hybridizing populations. Ultimately, the impact of fungal meiotic drive in population divergence and speciation might depend on the actual abundance of spore killers in natural populations, as well as the specificities of the life cycle and population dynamics of each system.
POPULATION DYNAMICS OF SPORE KILLERS
Due to a transmission advantage at the gene level, meiotic drivers can invade a population even when they lower the fitness of their host (Burt and Trivers Citation2009). The study of this counterintuitive phenomenon has opened a fruitful area of research aimed at understanding and predicting the dynamics of these selfish genes in animals and plants, with a growing body of both theoretical and empirical work. Early studies on meiotic drive in mice were paralleled by theoretical population genetics (e.g., Lewontin and Dunn Citation1960), and since then theoretical studies on meiotic drive have multiplied and diversified (e.g., demographic model, Marshall Citation2009; spatial models, Rode et al. Citation2020), often with strong connections to empirical systems and showing impressive predictive powers under controlled conditions (e.g., Fishman and Kelly Citation2015; Hall and Dawe Citation2018). In comparison with meiotic drivers in plants and animals, however, the study of fungal spore killer’s population dynamics is still at an early stage: theoretical work is scarce and restricted to population genetics, and knowledge of dynamics in natural populations is fragmentary and limited to a few model species.
As of today, there are two population genetics studies of the condition for invasion, i.e., stable polymorphism or fixation of a spore killer in ascomycete fungi (Nauta and Hoekstra Citation1993; Martinossi-Allibert et al. Citation2021). Both studies take into account potential fitness costs associated with the killer, as well as killing advantage, the possibility for a net fitness benefit associated with killing. However, the two studies are quite distinct. The Nauta and Hoekstra (Citation1993) study, the first theoretical description of spore killers, is based on Sk-2 and Sk-3 in N. intermedia. It focuses on the creation of resistant genotypes through recombination between the poison and antidote functions of the killer. They show that a relative driver, such as a spore killer, has a selective advantage that is proportional to the amount of killing per generation, which is in turn dependent on the frequency of the killer itself in the population, assuming random mating. Working in an infinite population size framework, the authors conclude that invasion of a spore killer without additional fitness benefits is impossible, because the selective advantage that a single killer derives from killing in an infinite population is effectively null.
In the more recent study, Martinossi-Allibert et al. (Citation2021) describe the population genetics of a single-gene killer. They show that in a finite population framework, invasion is possible due to a non-negligible selective advantage, provided that populations are not too large or are fragmented ()
Figure 5. Simulation of the invasion probability of a spore killer as a function of population size.
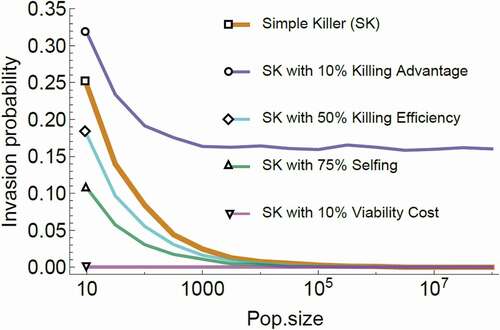
In summary, population genetics models predict that invasion of a spore killer should be possible by the simple action of killing, especially in small or fragmented populations (Martinossi-Allibert et al. Citation2021; but see Sweigart et al. Citation2019 for a verbal neutral theory of the evolution of spore killers). Fitness costs to the killer will prevent invasion () unless they are recessive or balanced by a compensation phenomenon (Nauta and Hoekstra Citation1993; Martinossi-Allibert et al. Citation2021).
Understanding the conditions that allow for coexistence between a meiotic driver and a nondriving allele are important, as meiotic drivers under stable polymorphism are much more likely to be observed (a meiotic driver can go to fixation rapidly and the driving phenotype subsequently disappears; Lindholm et al. Citation2016). Both spore killer models show that if a spore killer is 100% efficient (no sensitive spore exposed to killing survives), there can be no coexistence between killer and sensitive alleles. In this case, if a killer can invade, it should subsequently go to fixation (Nauta and Hoekstra Citation1993; Martinossi-Allibert et al. Citation2021). Coexistence between killer and sensitive alleles becomes possible if (i) killing efficiency is not complete, (ii) a resistant genotype evolves through recombination between the toxin and antidote functions (Nauta and Hoekstra Citation1993), or (iii) selfing of the host reduces the transmission rate of the killer (Martinossi-Allibert et al. Citation2021). In all cases, an additional necessary condition is the existence of fitness differences between the genotypes due to fitness costs, killing advantage, or both, resulting in some form of heterozygote advantage that favors invasion of the killer but prevents its fixation. Note that in ascomycetes that do not have an extended vegetative dikaryotic stage, such recessive effects can only arise during the very short-lived diploid stage.
Recombination between the killing and antidote functions leading to a nonkilling resistant genotype is unlikely in most known spore killing systems (see Genomic Architecture of Spore Killers). However, resistance may appear in a number of other ways (see Mechanisms of Resistance) and may be common in N. intermedia and S. pombe. The role of selfing, or inbreeding in heterothallic species, could be crucial in determining the fate of spore killers; coevolution between reproductive strategy and costly selfish genetic elements is supported in several other systems (e.g., mate choice in the stalk-eyed fly, Wilkinson et al. Citation1998; breeding system in plants, Burt and Trivers Citation1998). Selfing or inbreeding could thus be an efficient strategy to counteract the invasion of spore killers across fungi.
From only two theoretical studies, we still have a limited understanding of spore killer dynamics. However, both studies agree that spore killers present unique dynamics as compared with other types of meiotic drivers and thus deserve more attention, since their behavior cannot simply be extrapolated from other systems. A productive step forward would be to start systematic empirical testing of theoretical predictions. Some observations on frequencies of Spok genes in P. anserina seem to match the predictions that selfing rate of the host and fitness costs associated with the killer can become limiting factors over killing efficiency (Martinossi-Allibert et al. Citation2021; Vogan et al. Citation2021). This suggests that the diversity and flexibility of fungal mating strategies may indeed play a role in shaping the landscape of spore killers across species. There is one additional observation consistent with the model prediction stating that coexistence requires incomplete killing or resistance: in N. sitophila, the Spk-1 killer is fixed, absent, or polymorphic in three different clades, respectively; in the polymorphic clade, resistance to killing is present (Svedberg et al. Citation2021).
Population dynamics of spore killers is still a very young field, with an incomplete theoretical frame and only fragmented knowledge from a few model systems. Nevertheless, as recent empirical efforts have advanced our knowledge of spore killing in several ascomycetes, and the field of meiotic drive research is very much alive with the perspective of engineering synthetic drive, there is a will and a need to push forward. Different aspects of the theory will have to be explored, including demographic effects of spore killing, spatial models of spore killer invasion, the evolution of different types of resistance to killing, and more. All models will have to rely on ecological data from natural populations, outcrossing rates, demography, survival rates, and migration rates, which should be a focus of future collection efforts.
SPORE KILLERS AND SYNTHETIC DRIVE
Using naturally occurring meiotic drive to control populations was suggested almost as early as the first observations of sex-ratio distortion, which can have dramatic effects on the demography of species with separate sexes (Craig et al. Citation1960). The idea of controlling demography, which offered perspectives such as managing the malaria vector Aedes aegypti, was soon tested on caged populations of mosquitoes (Curtis et al. Citation1976), but the method lacked flexibility due to the limitation of genetic manipulation techniques available at the time (Burt and Crisanti Citation2018). The advent of more refined genetic engineering methods opened the possibility to engineer synthetic drivers as well as to link a gene of interest designated as “cargo” to a driver, natural or synthetic (e.g., Hammond et al. Citation2016). These technical advances permit more nuanced approaches, not limited to lowering the demography of the target population but allowing the introduction of genes of interest regardless of their fitness effects. They also allow for the implementation of rudimentary safety mechanisms to lower the risks of spreading to nontarget populations or organisms (Esvelt et al. Citation2014; Champer et al. Citation2016; Burt and Crisanti Citation2018; Wedell et al. Citation2019; Rode et al. Citation2020). However, despite potential applications with huge impact on public health (e.g., malaria vector control) and conservation (control of invasive species), implementation in natural populations remains to be seen, because of perceived risks from the scientific community as well as the public (Long et al. Citation2020; Rode et al. Citation2019; Wedell et al. Citation2019).
A key aspect in engineering both efficient and safe synthetic drivers is to understand and accurately predict the dynamics of the driving element once released in a natural environment (Backus and Delborne Citation2019; Wedell et al. Citation2019). This will require a detailed knowledge of fitness effects of the driver across environments and genetic backgrounds, as well as of the demography of the target population (Backus and Delborne Citation2019; Courtier-Orgogozo et al. Citation2020). For example, theoretical models show the key importance of spatial structure in the target population, which can allow the evolution of resistance (Bull et al. Citation2019) or interact with and disrupt potential safety mechanisms (e.g., gene brake; Girardin et al. Citation2019). The challenge of applied uses of synthetic drive thus goes beyond genetic engineering and requires a leap forward in the predictive power of evolutionary biology. It may take years of careful research before the safe introduction of a single synthetic drive in a natural population is made possible, and similar efforts may be needed to tailor the method to the specificities of each target population.
Spore killers may represent an opportunity to use a natural drive system to control populations of ascomycete fungi, for example by reducing virulence of crop pathogens such as Fusarium graminearum. As in other applications of synthetic drive, the potential benefits are large, including better crop yields and reduced use of fungicides (Gardiner et al. Citation2020). This potential use has been demonstrated in vitro, with the successful introduction of Spok1 (initially found in P. comata) in laboratory populations of F. graminearum (Gardiner et al. Citation2020). Thus, as with other types of synthetic drivers, there is an experimental proof of concept that harnessing spore killers for applied uses is feasible (Gardiner et al. Citation2020), but much more work is needed before population dynamics can be predicted (see Population Dynamics of Spore Killers) and an actual safe application is possible. Nevertheless, with the knowledge available today, we can examine the pros and cons of using naturally occurring spore killers as a basis for the development of synthetic drive.
A first argument in favor of using spore killers to develop synthetic drive is that there are already several well-known genetic elements in several species of ascomycetes that are potential candidates to choose from. Most represent short genetic sequences (see Genetic Architecture of Spore Killers), which makes them easily transferable across species (e.g., Nuckolls et al. Citation2020). Second, there are several potential mechanisms of host resistance in ascomycetes, including the evolution of suppressors and host genome defense mechanisms, that could be seen as useful features, since they have the potential to be co-opted as safety mechanisms to “recall” a synthetic driver if needed (as suggested by Gardiner et al. Citation2020). At the same time, such defense mechanisms could also appear as a downside, since they could make the introduction of synthetic drivers difficult or impossible altogether. One additional potential flaw in the idea of using synthetic drivers at all in ascomycetes is the fact that these selfish genetic elements rely on outcrossing to spread in the host population. This requirement may become problematic if asexual reproduction, selfing, or even inbreeding are frequent in populations of ascomycete fungi (see Population Dynamics of Spore Killers). On this last point we can only speculate, but the prevalence of spore killers in several species of ascomycetes shows that they are indeed able to invade; yet, the pace of that invasion is unknown and may be very slow if outcrossing events are rare in natural conditions. The fact that the spore killer introduced in laboratory populations of F. graminearum by Gardiner et al. (Citation2020) was able to spread within a few generations, even though the host was allowed to freely choose its mode of reproduction, is encouraging. In addition, several studies suggest that fungal pathogens in particular may increase outcrossing frequency due to the need of rapid coevolution with their host (Kaltz and Shykoff Citation1999; Laine et al. Citation2019). Finally, spore killers may naturally be candidates for being engineered as threshold dependent drivers, a type of driver that presents lower risks of invading nontarget populations because they require a minimum frequency in the population to start driving efficiently (Marshall and Akbari Citation2018; Champer et al. Citation2020; Girardin et al. Citation2019). As shown by population genetics models (Nauta and Hoekstra Citation1993; Martinossi-Allibert et al. Citation2021), spore killers have a naturally low selective advantage at low frequency, so a small associated fitness cost would be enough to make them threshold dependent drivers.
In conclusion, spore killers have several features that could make them a useful basis for the engineering of synthetic drivers in ascomycetes, which could in turn prove useful for eliminating plant pathogens or reducing their virulence. As for other types of meiotic drivers, natural or synthetic, their population dynamics need to be much better understood before applied use in natural populations should be envisioned.
HOW COMMON IS SPORE KILLING?
The diversity of the identified spore killer genes is high, with each spore killing gene having a generally limited phylogenetic distribution. In the three most studied fungal genera (Neurospora, Podospora, and Schizosaccharomyces), five distinct genes or gene families have been identified, and of these, two (Spok and wtf) show high within-family diversification with corresponding functional divergence in poison-antidote specificity. All of these gene families appear to be species or genus specific, with the exception of the Spok family, where homologous sequences can be found throughout Pezizomycotina (Grognet et al. Citation2014; Vogan et al. Citation2019). Even in the case of the Spok genes, it is still unclear whether these homologs have any spore killing activity, or whether this is a unique mutational event in Podospora that caused an existing gene to gain meiotic drive activity.
In any case, these results suggest that the potential for de novo evolution of spore killer genes is large. Neurospora, Podospora, and Schizosaccharomyces are all well-studied model systems for fungal biology, and the other species where spore killing has been observed (F. verticillioides, F. subglutinans, and B. maydis) are important plant pathogens. Spore killing was identified in these cases because different natural isolates were crossed to each other and either aberrant patterns of dead spores or segregation distortion of known markers were observed. Such crosses have only been done in a small number of species, and this fact, together with the high diversity and limited phylogenetic distribution of the known spore killer genes, makes it highly likely that the vast majority of spore killer genes remain unsampled.
There are several reasons why potential spore killers may not have been discovered, even in well-studied systems. Many important plant pathogens are not known to have a sexual cycle, and in others, sex is rare or dominated by selfing. In the case of obligate clonal species, spore killing should be unable to spread through a population, but genes that were active in a sexual ancestor may still remain functional. In the case of species where outcrossing is rare or difficult due to reduced fertility, researchers may have focused on other parts of the biology and missed it. One example of this is S. pombe, where spore killing was first described in 2017 (Hu et al. Citation2017; Nuckolls et al. Citation2017) despite decades of intense laboratory research. In this case, spore killing was not observed due to a combination of difficulty distinguishing dead from living spores under a microscope, the availability of only a small number of natural isolates, and severe reproductive barriers between these isolates. Interestingly, these reproductive barriers may in part be caused by the spore killer genes themselves.
It may be informative to compare this case with Saccharomyces cerevisiae, and other members of Saccharomycotina, which are model organisms for genetics and numerous other fields but which have no reported spore killers. The genomes of common laboratory strains of S. cerevisiae are well characterized and the presence of a single spore killer gene in these genomes would most likely have been identified, but this may not be the case for genes that are segregating in nature. Although many natural isolates are available, the types of experiments and procedures that could reveal the presence of spore killing appear to be rare. Some features of Saccharomyces biology may make them less likely to harbor meiotic drivers. For instance, large population sizes may make even small negative fitness effects of carrying extra spore killer genes be exposed to selection and relatively quickly purged. Additionally, S. cerevisiae outcrosses rarely as compared with other closely related yeast species (Niewenhuis and James Citation2016). On the other hand, some strains of S. cerevisiae and other yeasts contain so-called “killer plasmids,” which are also toxin-antidote systems that kill all yeast that do not carry the plasmid (Liu et al. Citation2015). This ensures segregation of the plasmid at both mitosis and meiosis, and it has similar evolutionary consequences and dynamics as spore killer genes (Boynton Citation2019). Thus, it is possible that spore killers are present in the Saccharomycotina, and future screening experiments, perhaps focused on species that outcross often, have the potential to identify such genetic elements.
All known spore killers have so far been found in ascomycete fungi. Is it possible that spore killing could also happen among other fungal clades? Ascomycete meiosis and sporogenesis takes place in a single cell (the ascus), where the four meiotic products will initially share a syncytium and then form individual spores by building cell walls around themselves. This creates a favorable environment for spore killing, where proteins generated from one meiotic product can easily diffuse throughout the ascus and any potential killing factor can reach a sensitive target. In basidiomycetes, meiosis and sporogenesis occur in a single cell as well; the basidium, and basidiospores are then formed through budding. This developmental program creates similar opportunities for spore killing, but it may also be more difficult to distinguish living from dead spores. Segregation distortion has been observed in basidiomycetes (Larraya et al. Citation2000; Chang and Jung Citation2008; Vogan and Xu Citation2014), but the mechanism behind this phenomenon is not yet known. It would be of great interest to know whether spore killing is found among basidiomycetes and other fungal clades, and whether this has any other consequences on their lifestyle and biology, such as rate of outcrossing, genome size, de novo gene evolution rate, etc.
PERSPECTIVES
It has been over 50 years since spore killing was first observed in fungi, but only in recent years has an understanding of the genetics behind the phenomenon emerged. The first spore killer gene to be described was the prion form of het-s in 2003 (Dalstra et al. Citation2003), and since 2014, four more genes or gene families causing spore killing have been identified. We now can start to try to find generalizable patterns that are common among these systems.
The most striking finding is that four out of the five identified genes causing spore killing are single genes, capable of inducing meiotic drive by themselves. This is in stark contrast to what is seen in other eukaryotes, where most well-studied meiotic drive elements are caused by multiple interacting genes that are located in large nonrecombining regions, or on sex chromosomes, where the presence of multiple interacting genes may be likely or cannot easily be excluded. Still, for several reasons, it may be premature to claim that this difference is a general pattern. One is that the number of described spore killer genes is still low, and although the majority of these are single-gene systems, we also have examples of multigene complexes with Sk-2 and Sk-3 in N. intermedia. It could be the case that both single-gene and multigene drivers occur, but that single-gene drivers escape detection in most circumstances. Indeed, a single gene is less likely to be linked to deleterious mutations than a multigene system in a region of low recombination, and it might therefore be expected to sweep to fixation more quickly, making it more difficult to detect unless its invasion is slowed down by other factors. As many fungi have the ability to spread clonally or are self-fertile, this may cause spore killers to remain polymorphic for longer, which makes them more likely to be detected. On top of this, unlike sperm killing meiotic drive elements in plants and animals that kill gametes, spore killers actually reduce the number of offspring, and this can lower the selective advantage of the spore killer and may also increase the time when a spore killer gene remains polymorphic. This model suggests a large undetected diversity of single-gene meiotic drivers in nonfungal systems, and it also suggests that multigene spore killers may be less likely to spread in fungi, due to the extra negative fitness of carrying linked deleterious mutations. In either case, this question can only be answered through further sampling.
The results presented here suggest that a large diversity of spore killers may be found in nature—and that they may not just be genetic oddities, instead serving as major drivers in fungal diversification, through the erection of reproductive barriers or in extreme cases such as S. pombe by severely reducing the capacity for outcrossing. Furthermore, they may be expected to affect genome evolution, in the shape of gene duplication, de novo gene evolution, and the accumulation of structural rearrangements and transposable elements.
We still have much to learn in regard to the molecular mechanisms underlying the spore killing phenomenon, both in terms of the killing of sensitive spores and the protection of nonsensitive ones. Further work on the population and interspecies dynamics of spore killing is also necessary, both in the field and in a controlled laboratory setting, and it will be of great interest to see whether lessons learned from natural spore killers can be applied when considering the possibility of population control through synthetic drivers. With the high pace of methodological development in affordable genome sequencing and through techniques such as CRISPR (clustered regularly interspaced short palindromic repeats), biologists who study nonmodel systems are in an increasingly better position to identify and study this phenomenon. Spore killers often have the advantage of being easily identifiable using standard microscopes, and we encourage fungal biologists to be aware of the phenomenon and keep an eye out for these distinct patterns of dead and living spores.
GLOSSARY
Dikaryon: The condition of having two nuclear types per cell compartment. For example, Podospora species and N. tetrasperma have dikaryotic ascospores.
Enhancer: Genetic elements that increase the transmission advantage of a meiotic driver.
Heterothallism: Reproductive system where mating is only possible between individuals carrying different alleles in a mating type locus. It prevents haploid selfing.
Homothallism: Reproductive system where an individual is universally compatible with all individuals in the population, including itself. It allows haploid selfing.
Killing advantage: Attenuation of fitness cost imposed by a gamete killing/spore killing driver. For example, production of additional meiotic products to replace the killed ones, or any other fitness benefit for surviving ones.
Meiotic drivers: Selfish genetic elements that have the feature of being overrepresented among meiotic products, leading to non-Mendelian segregation patterns.
Monokaryon: The condition of having a single nuclear type per cell compartment.
Pseudohomothallism: Reproductive system derived from heterothallism where nuclei of different mating types are packaged together in sexual spores, leading to dikaryotic, self-compatible mycelia. It facilitates diploid selfing but does not allow haploid selfing.
Resistance: The phenotype of not being affected by a driving mechanism, while also not driving. Suppressors may confer partial or total resistance.
Suppressor: Genetic elements that reduce the transmission advantage of a meiotic driver.
DISCLOSURE STATEMENT
No potential conflict of interest was reported by the author(s).
Additional information
Funding
LITERATURE CITED
- Backus GA, Delborne JA. 2019. Threshold-dependent gene drives in the wild: spread, controllability, and ecological uncertainty. BioScience. 69(11):900–1007. doi:https://doi.org/10.1093/biosci/biz098
- Bernet J. 1965. Mode d’action des gènes de “barrage” et relation entre l’incompatibilité cellulaire et l’incompatibilité sexuelle chez “Podospora anserina” [doctoral dissertation]. Paris (France): University of Paris.
- Billiard S, López-Villavicencio M, Hood ME, Giraud T. 2012. Sex, outcrossing and mating types: unsolved questions in fungi and beyond. J Evol Biol. 25(6):1020–1038. doi:https://doi.org/10.1111/j.1420-9101.2012.02495.x.
- Boucher C, Nguyen T-S, Silar P. 2017. Species delimitation in the Podospora anserina/P. pauciseta/P. comata species complex (Sordariales). Cryptogam Mycol. 38(4):485–506. doi:https://doi.org/10.7872/crym/v38.iss4.2017.485.
- Bowen NJ, Jordan IK, Epstein JA, Wood V, Levin HL. 2003. Retrotransposons and their recognition of Pol II promoters: a comprehensive survey of the transposable elements from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 13(9):1984–1997. doi:https://doi.org/10.1101/gr.1191603.
- Boynton PJ. 2019. The ecology of killer yeasts: interference competition in natural habitats. Yeast. 36(8):473–485. doi:https://doi.org/10.1002/yea.3398.
- Bravo Núñez MA, Nuckolls NL, Zanders SE. 2018. Genetic villains: killer meiotic drivers. Trends Genet. 34(6):424–433. doi:https://doi.org/10.1016/j.tig.2018.02.003.
- Bravo Núñez MA, Sabbarini IM, Eickbush MT, Liang Y, Lange JJ, Kent AM, Zanders SE. 2020a. Dramatically diverse Schizosaccharomyces pombe wtf meiotic drivers all display high gamete-killing efficiency. PLoS Genet. 16(2):e1008350. doi:https://doi.org/10.1371/journal.pgen.1008350.
- Bravo Núñez MA, Sabbarini IM, Eide LE, Unckless RL, Zanders SE. 2020b. Atypical meiosis can be adaptive in outcrossed due to meiotic drivers. eLife. 9. doi:https://doi.org/10.7554/eLife.57936.
- Bronson CR. 1988. Ascospore abortion in crosses of Cochliobolus heterostrophus heterozygous for the virulence locus Tox1. Genome. 30(1):12–18. doi:https://doi.org/10.1139/g88-003.
- Bronson CR. 1990. Genetic control and distorted segregation of T-toxin production in field isolates of Cochliobolus heterostrophus. Phytopathology. 80(9):819. doi:https://doi.org/10.1094/Phyto-80-819.
- Bull JJ, Remien CH, Gomulkiewicz R, Krone SM. 2019. Spatial structure undermines parasite suppression by gene drive cargo. PeerJ. 7:e7921. doi:https://doi.org/10.7717/peerj.7921.
- Burt A, Crisanti A. 2018. Gene drive: evolved and synthetic. ACS Chem Biol. 13(2):343–346. doi:https://doi.org/10.1021/acschembio.7b01031.
- Burt A, Trivers R. 1998. Selfish DNA and breeding system in flowering plants. Proc R Soc Lond B. 265(1391):141–146. doi:https://doi.org/10.1098/rspb.1998.0275.
- Burt A, Trivers R. 2009. Genes in conflict: the biology of selfish genetic elements. Cambridge (MA): Harvard University Press. 612 p.
- Cambareri E, Jensen B, Schabtach E, Selker E. 1989. Repeat-induced G-C to A-T mutations in Neurospora. Science. 244(4912):1571–1575. doi:https://doi.org/10.1126/science.2544994.
- Campbell JL, Turner BC. 1987. Recombination block in the spore killer region of Neurospora. Genome. 29(1):129–135. doi:https://doi.org/10.1139/g87-022.
- Catalanotto C, Nolan T, Cogoni C. 2006. Homology effects in Neurospora crassa. FEMS Microbiol Lett. 254(2):182–189. doi:https://doi.org/10.1111/j.1574-6968.2005.00037.x.
- Cerutti H, Casas-Mollano JA. 2006. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 50(2):81–99. doi:https://doi.org/10.1007/s00294-006-0078-x.
- Champer J, Buchman A, Akbari OS. 2016. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet. 17(3):146–159. doi:https://doi.org/10.1038/nrg.2015.34.
- Champer J, Lee YL, Yang E, Liu C, Clark AG, Messer PW. 2020. A toxin-antidote CRISPR gene drive system for regional population modification. Nature Communications. 11:1082. doi:https://doi.org/10.1101/628354.
- Chang SW, Jung G. 2008. The first linkage map of the plant-pathogenic basidiomycete Typhula ishikariensis. Genome. 51(2):128–136. doi:https://doi.org/10.1139/G07-097.
- Charron Y, Willert J, Lipkowitz B, Kusecek B, Herrmann BG, Bauer H. 2019. Two isoforms of the RAC-specific guanine nucleotide exchange factor TIAM2 act oppositely on transmission ratio distortion by the mouse t-haplotype. PLoS Genet. 15(2):e1007964. doi:https://doi.org/10.1371/journal.pgen.1007964.
- Choi J, Kim K, Jeon J, Wu J, Song H, Asiegbu F O, Lee Y. 2014. funRNA: a fungi-centered genomics platform for genes encoding key components of RNAi. BMC Genomics. 15:S14. doi:https://doi.org/10.1186/1471-2164-15-S9-S14.
- Clutterbuck AJ. 2011. Genomic evidence of repeat-induced point mutation (RIP) in filamentous ascomycetes. Fungal Genet Biol. 48:306–326.
- Coleman JJ, et al. 2009. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5(8):e1000618. doi:https://doi.org/10.1371/journal.pgen.1000618.
- Courret C, Chang C-H, Wei KH-C, Montchamp-Moreau C, Larracuente AM. 2019. Meiotic drive mechanisms: lessons from. Proc Biol Sci. 286:20191430.
- Courtier-Orgogozo V, Danchin A, Gouyon P-H, Boëte C. 2020. Evaluating the probability of CRISPR-based gene drive contaminating another species. Evol Appl. 13(8):1888–1905. doi:https://doi.org/10.1111/eva.12939.
- Coustou V, Deleu C, Saupe S, Begueret J. 1997. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci U S A. 94(18):9773–9778. doi:https://doi.org/10.1073/pnas.94.18.9773.
- Coyne JA, Orr HA. 2004. Speciation. Sunderland (MA): Sinauer Associates. 545 p.
- Craig GB Jr, Hickey WA, Vandehey RC. 1960. An inherited male-producing factor in Aedes aegypti. Science. 132(3443):1887–1889. doi:https://doi.org/10.1126/science.132.3443.1887.
- Crespi B, Nosil P. 2013. Conflictual speciation: species formation via genomic conflict. Trends Ecol Evol. 28(1):48–57. doi:https://doi.org/10.1016/j.tree.2012.08.015.
- Crow JF. 1991b. Why is Mendelian segregation so exact? Bioessays. 13(6):305–312. doi:https://doi.org/10.1002/bies.950130609.
- Curtis CF, Grover KK, Suguna SG, Uppal DK, Dietz K, Agarwal HV, Kazmi SJ. 1976. Comparative field cage tests of the population suppressing efficiency of three genetic control systems for Aedes aegypti. Heredity. 36(1):11–29. doi:https://doi.org/10.1038/hdy.1976.2.
- Dalstra HJP, Swart K, Debets AJM, Saupe SJ, Hoekstra RF. 2003. Sexual transmission of the [Het-S] prion leads to meiotic drive in Podospora anserina. Proc Natl Acad Sci U S A. 100(11):6616–6621. doi:https://doi.org/10.1073/pnas.1030058100.
- Debets AJM, Dalstra HJP, Slakhorst M, Koopmanschap B, Hoekstra RF, Saupe SJ. 2012. High natural prevalence of a fungal prion. Proc Natl Acad Sci U S A. 109(26):10432–10437. doi:https://doi.org/10.1073/pnas.1205333109.
- Eickbush MT, Young JM, Zanders SE. 2019. Killer meiotic drive and dynamic evolution of the wtf gene family. Mol Biol Evol. 36:1201–1214.
- Esvelt KM, Smidler AL, Catteruccia F, Church GM. 2014. Concerning RNA-guided gene drives for the alteration of wild populations. eLife. 3. doi:https://doi.org/10.7554/eLife.03401.
- Fishman L, Kelly JK. 2015. Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution. 69(5):1208–1218. doi:https://doi.org/10.1111/evo.12661.
- Frank SA. 1991a. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution. 45:262–267.
- Frank SA. 1991b. Haldane’s rule: a defense of the meiotic drive theory. Evolution. 45(7):1714–1717. doi:https://doi.org/10.1111/j.1558-5646.1991.tb02678.x.
- Gardiner DM, Rusu A, Barrett L, Hunter GC, Kazan K. 2020. Can natural gene drives be part of future fungal pathogen control strategies in plants? New Phytol. 228(4):1431–1439. doi:https://doi.org/10.1111/nph.16779.
- Girardin L, Calvez V, Débarre F. 2019. Catch me if you can: a spatial model for a brake-driven gene drive reversal. Bull Math Biol. 81(12):5054–5088. doi:https://doi.org/10.1007/s11538-019-00668-z.
- Graïa F, Lespinet O, Rimbault B, Dequard-Chablat M, Coppin E, Picard M. 2001. Genome quality control: RIP (repeat-induced point mutation) comes to Podospora. Mol Microbiol. 40(3):586–595. doi:https://doi.org/10.1046/j.1365-2958.2001.02367.x.
- Grognet P, Lalucque H, Malagnac F, Silar P. 2014. Genes that bias Mendelian segregation. PLoS Genet. 10(5):e1004387. doi:https://doi.org/10.1371/journal.pgen.1004387.
- Hall DW, Dawe RK. 2018. Modeling the evolution of female meiotic drive in Maize. G3 8(1):123–130. doi:https://doi.org/10.1534/g3.117.300073.
- Hamann A, Osiewacz HD. 2004. Genetic analysis of spore killing in the filamentous ascomycete Podospora anserina. Fungal Genet Biol. 41(12):1088–1098. doi:https://doi.org/10.1016/j.fgb.2004.08.008.
- Hammond A, et al. 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 34(1):78–83. doi:https://doi.org/10.1038/nbt.3439.
- Hammond TM, Rehard DG, Xiao H, Shiu PKT. 2012. Molecular dissection of Neurospora spore killer meiotic drive elements. Proc Natl Acad Sci U S A. 109(30):12093–12098. doi:https://doi.org/10.1073/pnas.1203267109.
- Hansen KR, Burns G, Mata J, Volpe TA, Martienssen RA, Bähler J, Thon G. 2005. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol Cell Biol. 25(2):590–601. doi:https://doi.org/10.1128/MCB.25.2.590-601.2005.
- Hartl DL. 1972. Population dynamics of sperm and pollen killers. Theor Appl Genet. 42(2):81–88. doi:https://doi.org/10.1007/BF00277948.
- Harvey AM, Rehard DG, Groskreutz KM, Kuntz DR, Sharp KJ, Shiu PKT, Hammond TM. 2014. A critical component of meiotic drive in Neurospora is located near a chromosome rearrangement. Genetics. 197(4):1165–1174. doi:https://doi.org/10.1534/genetics.114.167007.
- Hernández JFL, Helston RM, Lange JJ, Blake Billmyre R, Schaffner SH, Eickbush MT, McCroskey S, Zanders SE. 2021. Diverse mating phenotypes impact the spread of wtf meiotic drivers in S. pombe. bioRxiv. doi:https://doi.org/10.1101/2021.05.28.446231.
- Horns F, Petit E, Yockteng R, Hood ME. 2012. Patterns of repeat-induced point mutation in transposable elements of basidiomycete fungi. Genome Biol Evol. 4(3):240–247. doi:https://doi.org/10.1093/gbe/evs005.
- Hu W, Jiang Z-D, Suo F, Zheng J-X, He W-Z, Du -L-L. 2017. A large gene family in fission yeast encodes spore killers that subvert Mendel’s law. eLife. 6. doi:https://doi.org/10.7554/eLife.26057.
- Hurst LD, Pomiankowski A. 1991. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics. 129(2):603. doi:https://doi.org/10.1093/genetics/129.2.603.
- Jaenike J. 2001. Sex chromosome meiotic drive. Annu Rev Ecol Syst. 32(1):25–49. doi:https://doi.org/10.1146/annurev.ecolsys.32.081501.113958.
- Kaltz O, Shykoff JA. 1999. Selfing versus outcrossing propensity of the fungal pathogen Microbotryum violaceum across Silene latifolia host plants. J Evol Biol. 12(2):340–349. doi:https://doi.org/10.1046/j.1420-9101.1999.00014.x.
- Kathariou S, Spieth PT. 1982. Spore killer polymorphism in Fusarium moniliforme. Genetics. 102(1):19–24. doi:https://doi.org/10.1093/genetics/102.1.19.
- Laine A-L, Barrès B, Numminen E, Siren JP. 2019. Variable opportunities for outcrossing result in hotspots of novel genetic variation in a pathogen metapopulation. eLife. 8:e47091.
- Larraya LM, Pérez G, Ritter E, Pisabarro AG, Ramírez L. 2000. Genetic linkage map of the edible basidiomycete Pleurotus ostreatus. Appl Environ Microbiol. 66(12):5290–5300. doi:https://doi.org/10.1128/AEM.66.12.5290-5300.2000.
- Lewontin RC, Dunn LC. 1960. The evolutionary dynamics of a polymorphism in the house mouse. Genetics. 45(6):705–722. doi:https://doi.org/10.1093/genetics/45.6.705.
- Lindholm AK, et al. 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol Evol. 31(4):315–326. doi:https://doi.org/10.1016/j.tree.2016.02.001.
- Liu G-L, Chi Z, Wang G-Y, Wang Z-P, Li Y, Chi Z-M. 2015. Yeast killer toxins, molecular mechanisms of their action and their applications. Crit Rev Biotechnol. 35(2):222–234. doi:https://doi.org/10.3109/07388551.2013.833582.
- Long KC, et al. 2020. Core commitments for field trials of gene drive organisms. Science. 370(6523):1417–1419. doi:https://doi.org/10.1126/science.abd1908.
- Lutzoni F, et al. 2018. Contemporaneous radiations of fungi and plants linked to symbiosis. Nat Commun. 9(1):5451. doi:https://doi.org/10.1038/s41467-018-07849-9.
- Lyttle TW. 1991. Segregation distorters. Annu Rev Genet. 25(1):511–557. doi:https://doi.org/10.1146/annurev.ge.25.120191.002455.
- Ma L-J, Geiser DM, Proctor RH, Rooney AP, O’Donnell K, Trail F, Gardiner DM, Manners JM, Kazan K. 2013. Fusarium pathogenomics. Annu Rev Microbiol. 67(1):399–416. doi:https://doi.org/10.1146/annurev-micro-092412-155650.
- Marshall JM. 2009. The effect of gene drive on containment of transgenic mosquitoes. J Theor Biol. 258(2):250–265. doi:https://doi.org/10.1016/j.jtbi.2009.01.031.
- Marshall JM, Akbari OS. 2018. Can CRISPR-based gene drive be confined in the wild? A question for molecular and population biology. ACS Chem Biol. 13:424–430. doi:https://doi.org/10.1021/acschembio.7b00923.
- Martinossi-Allibert I, Veller C, Ament-Velásquez SL, Vogan AA, Rueffler C, Johannesson H. 2021. Invasion and maintenance of meiotic drivers in populations of ascomycete fungi. Evolution. 75(5):1150–1169. doi:https://doi.org/10.1111/evo.14214.
- Meiklejohn CD, et al. 2018. Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. eLife. 7. doi:https://doi.org/10.7554/eLife.35468.
- Milgroom MG, Smith ML, Drott MT, Nuss DL. 2018. Balancing selection at nonself recognition loci in the chestnut blight fungus, Cryphonectria parasitica, demonstrated by trans-species polymorphisms, positive selection, and even allele frequencies. Heredity. 121(6):511–523. doi:https://doi.org/10.1038/s41437-018-0060-7.
- Nakayashiki H, Kadotani N, Mayama S. 2006. Evolution and diversification of RNA silencing proteins in fungi. J Mol Evol. 63(1):127–135. doi:https://doi.org/10.1007/s00239-005-0257-2.
- Nauta MJ, Hoekstra RF. 1993. Evolutionary dynamics of spore killers. Genetics. 135(3):923–930. doi:https://doi.org/10.1093/genetics/135.3.923.
- Nguyen D, Peona V, Unneberg P, Suh A, Jern P, Johannesson H. 2020. Transposon- and genome dynamics in the fungal genus Neurospora: insights from nearly gapless genome assemblies. bioRxiv. doi:https://doi.org/10.1101/2020.09.27.311811.
- Nieuwenhuis BPS, James TY. 2016. The frequency of sex in fungi. Philos Trans R Soc B. 371(1706):20150540. doi:https://doi.org/10.1098/rstb.2015.0540.
- Nuckolls NL, et al. 2020. The meiotic driver utilizes controlled protein aggregation to generate selective cell death. eLife. 9. doi:https://doi.org/10.7554/eLife.55694.
- Nuckolls NL, Bravo Núñez MA, Eickbush MT, Young JM, Lange JJ, Yu JS, Smith GR, Jaspersen SL, Malik HS, Zanders SE. 2017. Wtf genes are prolific dual poison-antidote meiotic drivers. eLife. 6:e26033.
- Núñez MAB, Lange JJ, Zanders SE. 2018. A suppressor of a wtf poison-antidote meiotic driver acts via mimicry of the driver’s antidote. PLoS Genet. 14(11):e1007836. doi:https://doi.org/10.1371/journal.pgen.1007836.
- O’Donnell K, Nirenberg HI, Aoki T, Cigelnik E. 2000. A Multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 41(1):61–78. doi:https://doi.org/10.1007/bf02464387.
- Orr A. 1996. Dobzhansky, Bateson, and the genetics of speciation. Genetics. 144:1331–1335. doi:https://doi.org/10.1093/genetics/144.4.1331.
- Padieu E, Bernet J. 1967. Mode of action of the genes responsible for abortion of certain products of meiosis in the Ascomycete, Podospora anserina. C R Acad Hebd Seances Acad Sci D. 264:2300–2303.
- Perkins DD. 1974. The manifestation of chromosome rearrangements in unordered asci of Neurospora. Genetics. 77(3):459–489. doi:https://doi.org/10.1093/genetics/77.3.459.
- Pinan-Lucarré B, Paoletti M, Clavé C. 2007. Cell death by incompatibility in the fungus Podospora. Semin Cancer Biol. 17(2):101–111. doi:https://doi.org/10.1016/j.semcancer.2006.11.009.
- Pyle J, Patel T, Merrill B, Nsokoshi C, McCall M, Proctor RH, Brown DW, Hammond TM. 2016. A meiotic drive element in the maize pathogen Fusarium verticillioides is located within a 102 kb region of chromosome V. G3. 6(8):2543–2552. doi:https://doi.org/10.1534/g3.116.029728.
- Raju NB, Metzenberg RL, Shiu PKT. 2007. Neurospora spore killers Sk-2 and Sk-3 suppress meiotic silencing by unpaired DNA. Genetics. 176(1):43–52. doi:https://doi.org/10.1534/genetics.106.069161.
- Raju N, Perkins D. 1994. Diverse programs of ascus development in pseudohomothallic species of Neurospora, Gelasinospora, and Podospora. Dev Genet. 15(1):104–118. doi:https://doi.org/10.1002/dvg.1020150111.
- Reis M, Vieira CP, Lata R, Posnien N, Vieira J. 2018. Origin and consequences of chromosomal inversions in the virilis group of Drosophila. Genome Biol Evol. 10(12):3152–3166. doi:https://doi.org/10.1093/gbe/evy239.
- Rhoades MM. 1942. Preferential segregation in maize. Genetics. 27(4):395–407. doi:https://doi.org/10.1093/genetics/27.4.395.
- Rhoades NA, et al. 2019. Identification of rfk-1, a meiotic driver undergoing RNA editing in Neurospora. Genetics. 212(1):93–110. doi:https://doi.org/10.1534/genetics.119.302122.
- Riek R, Saupe SJ. 2016. The HET-S/s prion motif in the control of programmed cell death. Cold Spring Harb Perspect Biol. 8(9):a023515. doi:https://doi.org/10.1101/cshperspect.a023515.
- Rode NO, Courtier-Orgogozo V, Débarre F. 2020. Can a population targeted by a CRISPR-based homing gene drive be rescued? G3. 10(9):3403–3415. doi:https://doi.org/10.1534/g3.120.401484.
- Rode NO, Estoup A, Bourguet D, Courtier-Orgogozo V, Débarre F. 2019. Population management using gene drive: molecular design, models of spread dynamics and assessment of ecological risks. Conserv Genet. 20(4):671–690. doi:https://doi.org/10.1007/s10592-019-01165-5.
- Shiu PK, Raju NB, Zickler D, Metzenberg RL. 2001. Meiotic silencing by unpaired DNA. Cell. 107(7):905–916. doi:https://doi.org/10.1016/S0092-8674(01)00609-2.
- Sidhu GS. 1984. Genetics of Gibberella fujikuroi V. Spore killer alleles in G. fujikuroi. J Hered. 75(3):237–238. doi:https://doi.org/10.1093/oxfordjournals.jhered.a109923.
- Son H, Min K, Lee J, Raju NB, Lee Y-W. 2011. Meiotic silencing in the homothallic fungus Gibberella zeae. Fungal Biol. 115(12):1290–1302. doi:https://doi.org/10.1016/j.funbio.2011.09.006.
- Summerell BA. 2019. Resolving Fusarium: current status of the genus. Annu Rev Phytopathol. 57(1):323–339. doi:https://doi.org/10.1146/annurev-phyto-082718-100204.
- Sun Y, Svedberg J, Hiltunen M, Corcoran P, Johannesson H. 2017. Large-scale suppression of recombination predates genomic rearrangements in Neurospora tetrasperma. Nat Commun. 8(1):1140. doi:https://doi.org/10.1038/s41467-017-01317-6.
- Svedberg J, et al. 2018. Convergent evolution of complex genomic rearrangements in two fungal meiotic drive elements. Nat Commun. 9(1):4242. doi:https://doi.org/10.1038/s41467-018-06562-x.
- Svedberg J, Vogan AA, Rhoades NA, Sarmarajeewa D, Jacobson DJ, Lascoux M, Hammond TM, Johannesson H. 2021. An introgressed gene causes meiotic drive in Neurospora sitophila. Proc Natl Acad Sci U S A. 118(17): e2026605118.
- Sweigart AL, Brandvain Y, Fishman L. 2019. Making a Murderer: the evolutionary framing of hybrid gamete-killers. Trends Genet. 35(4):245–252. doi:https://doi.org/10.1016/j.tig.2019.01.004.
- Taga M, Bronson CR, Yoder OC. 1985. Nonrandom abortion of ascospores containing alternate alleles at the Tox-1 locus of the fungal plant pathogen Cochliobolus heterostrophus. Can J Genet Cytol. 27:450–456. doi:https://doi.org/10.1139/g85-066.
- Turgeon BG, Baker SE. 2007. Genetic and genomic dissection of the Cochliobolus heterostrophus Tox1 locus controlling biosynthesis of the polyketide virulence factor T-toxin. Adv Genet. 57:219–261.
- Turgeon BG, Kodama M, Yang G, Rose MS, Lu SW, Yoder OC. 1995. Function and chromosomal location of the Cochliobolus heterostrophus TOX1 locus. Can J Bot. 73(S1):1071–1076. doi:https://doi.org/10.1139/b95-359.
- Turner BC. 2001. Geographic distribution of Neurospora spore killer strains and strains resistant to killing. Fungal Genet Biol. 32(2):93–104. doi:https://doi.org/10.1006/fgbi.2001.1253.
- Turner BC, Perkins DD. 1979. Spore killer, a chromosomal factor in Neurospora that kills meiotic products not containing it. Genetics. 93(3):587–606. doi:https://doi.org/10.1093/genetics/93.3.587.
- Turner BC, Perkins DD. 1991. Meiotic drive in Neurospora and other fungi. Am Nat. 137(3):416–429. doi:https://doi.org/10.1086/285174.
- Tusso S, Nieuwenhuis BPS, Sedlazeck FJ, Davey JW, Jeffares DC, Wolf JBW. 2019. Ancestral admixture is the main determinant of global biodiversity in fission yeast. Mol Biol Evol. 36:1975–1989.
- Van der Gaag M, Debets AJ, Oosterhof J, Slakhorst M, Thijssen JA, Hoekstra RF. 2000. Spore-killing meiotic drive factors in a natural population of the fungus Podospora anserina. Genetics. 156(2):593–605. doi:https://doi.org/10.1093/genetics/156.2.593.
- Vogan AA, et al. 2019. Combinations of Spok genes create multiple meiotic drivers in Podospora. eLife. 8. doi:https://doi.org/10.7554/eLife.46454.
- Vogan AA, Ament-Velásquez SL, Bastiaans E, Wallerman O, Saupe SJ, Suh A, Johannesson H. 2021. The Enterprise, a massive transposon carrying meiotic drive genes. Genome Res. 31:789–798. doi:https://doi.org/10.1101/gr.267609.120.
- Vogan AA, Xu J. 2014. Evidence for genetic incompatibilities associated with post-zygotic reproductive isolation in the human fungal pathogen Cryptococcus neoformans. Genome. 57(6):335–344. doi:https://doi.org/10.1139/gen-2014-0077.
- Vogan A, Svedberg J, Grudzinska-Sterno M, Johannesson H. 2020. Meiotic drive is associated with sexual incompatibility in Neurospora. bioRxiv. doi:https://doi.org/10.1101/2020.11.27.400796.
- Wedell N, Price TAR, Lindholm AK. 2019. Gene drive: progress and prospects. Proc R Soc B. 286(1917):20192709. doi:https://doi.org/10.1098/rspb.2019.2709.
- Wilkinson GS, Kahler H, Baker RH. 1998. Evolution of female mating preferences in stalk-eyed flies. Behav Ecol. 9(5):525–533. doi:https://doi.org/10.1093/beheco/9.5.525.
- Wright SI, Ness RW, Foxe JP, Barrett SCH. 2008. Genomic consequences of outcrossing and selfing in plants. Int J Plant Sci. 169(1):105–118. doi:https://doi.org/10.1086/523366.
- Zanders SE, Eickbush MT, Yu JS, Kang J-W, Fowler KR, Smith GR, Malik HS. 2014. Genome rearrangements and pervasive meiotic drive cause hybrid infertility in fission yeast. eLife. 3:e02630. doi:https://doi.org/10.7554/eLife.02630.
- Zanders SE, Unckless RL. 2019. Fertility costs of meiotic drivers. Curr Biol. 29:R512–R520.
